Figure 3. Tn7017 exploits distinct TniQ homologs for two different targeting pathways.
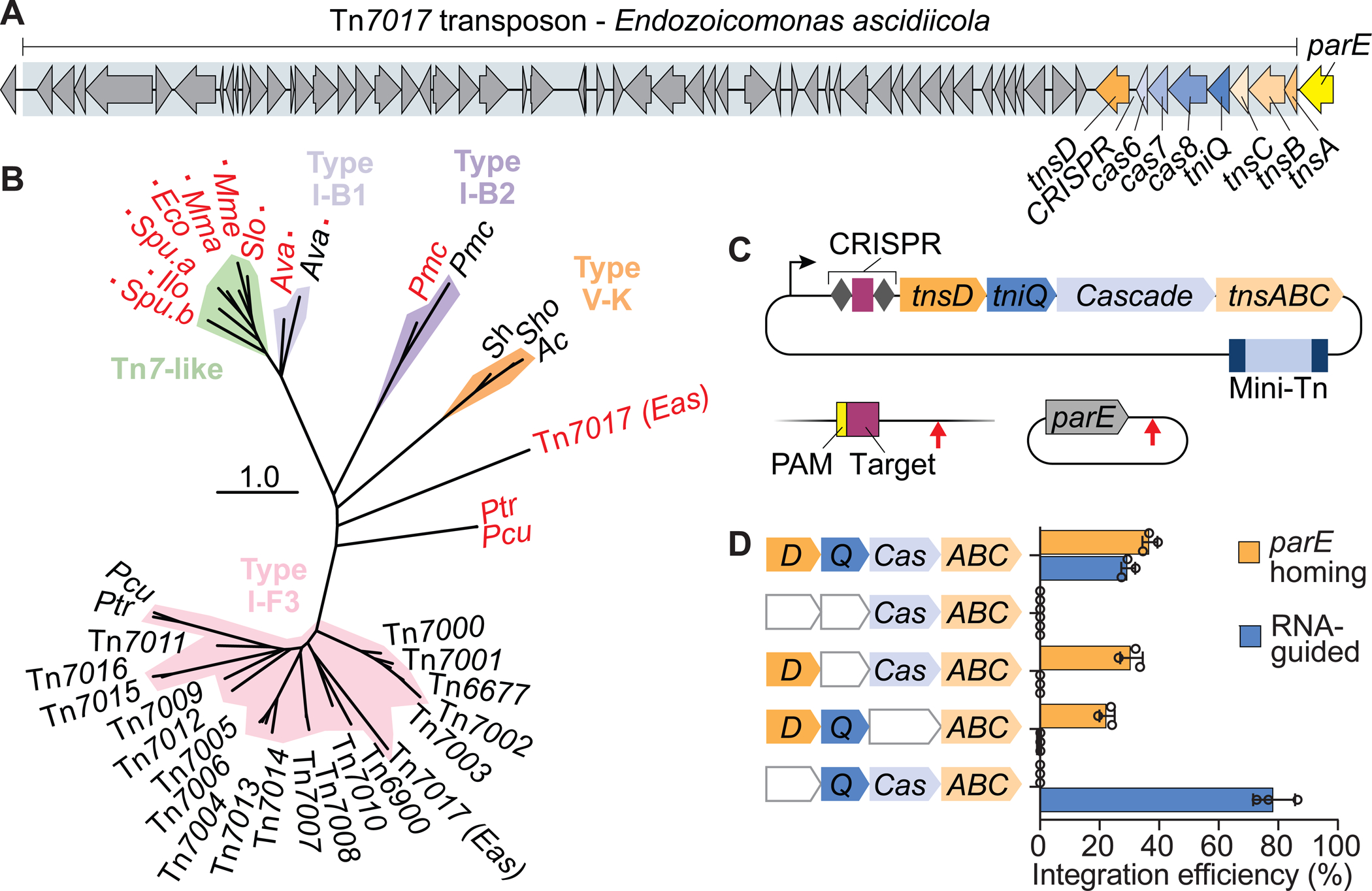
(A) Schematic representation of Tn7017 from E. ascidiicola, showing the presence of two distinct TniQ-family genes, labeled tniQ and tnsD. ‘TniQ’ and ‘TnsD’ are used to describe TniQ-family proteins involved in the RNA-dependent and RNA-independent targeting pathways, respectively. (B) Pruned phylogenetic tree of TniQ/TnsD family proteins based on tniQ domain (PF06527) alignment. Clades containing known Tn7-like transposons and CRISPR-Tn systems (I-B1, I-B2, V-K, and I-F3) are indicated. Black text indicates a TniQ protein, red text indicates a TnsD protein. A red square indicates that a TnsD domain (PF15978) was identified in the full-length protein. Also shown are TniQ and TnsD proteins from two Parashewanella species (Ptr and Pcu) described in Petassi et al. (2020), which are also predicted to facilitate RNA-guided transposition and RNA-independent parE homing, respectively. (C) Transposition assay design for simultaneous detection of DNA integration at a genomic target site (RNA-dependent) and a putative, plasmid-borne attachment site (RNA-independent). Arrows indicate the predicted integration sites relative to the target sites. (D) Integration efficiency for pTarget and the genomic target site, as measured by qPCR, under different gene deletion conditions. Data are shown as mean ± s.d. for n = 3 biologically independent samples. See also Figure S3.
