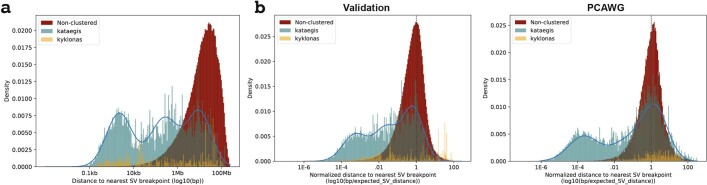
Extended Data Fig. 9. Kyklonas occur distally from structural breakpoints across three independent cohorts.
a, The distance to the nearest breakpoint for all kataegic mutations (teal), kyklonas (gold), and non-clustered mutations (red) across the three validation cohorts. b, Distances to the nearest SV breakpoints were normalized by calculating the expected distance a mutation would fall from a breakpoint given the number of breakpoints detected per chromosome and the overall length of the chromosome across the validation cohorts (left) and PCAWG (right). A value of 1 (dashed line) reflects a distance that one would expect based on the random placement of a mutation across the chromosome, whereas a value less than 1 reflects a mutation occurring closer than what is expected by random chance. The distributions of kataegic mutations were modelled using Gaussian mixture models (blue lines) with an automatic selection criterion for the number of components using the minimum Bayesian information criteria (BIC).