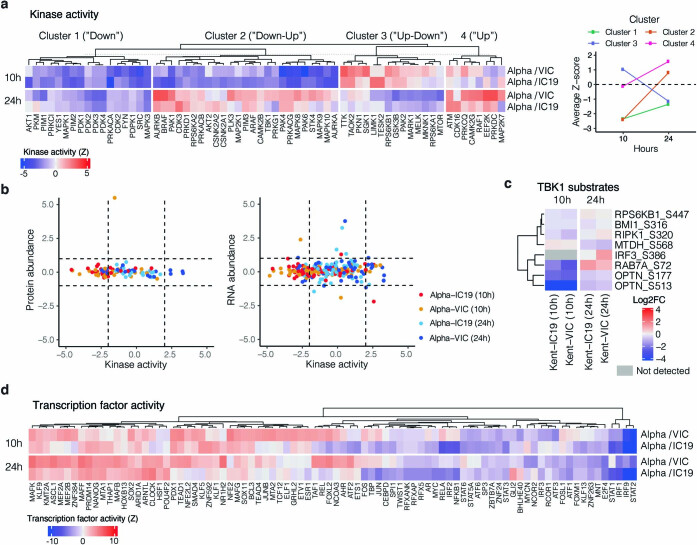
Extended Data Fig. 6. Kinase and transcription factor activity analysis.
a, Full kinase activity analysis of indicated contrasts with z-score>2. Kinases were separated using k-means clustering, which naturally reveals groups depicting kinases downregulated for the entire time course (“Down”), downregulated early and upregulated late (“Down-Up”), upregulated early and downregulated late (“Up-Down”), or upregulated or constant throughout the time course (“Up”). Panel on the right depicts the average Z-score for each distinct cluster per time point, collapsing across Alpha/VIC and Alpha/IC19 comparisons. b, Correlation between the calculated kinase activity Z-score and protein (left) or RNA (right) abundance log2FC for kinases with estimated activities in our dataset. Vertical dashed lines indicate kinase activity of ±2, horizontal dashed lines indicate protein log2FC of ±1. Colours represent comparisons between viruses and time points as indicated. c, Detected substrates known to be phosphorylated by TBK1. Log2FC of each phosphorylation site is depicted. Those not detected are indicated in grey. d, Transcription factor (TF) activities were estimated from the RNA-seq dataset using known TF-target gene interactions. Included are TFs with a NES>2.5. TF are clustered using ward hierarchical clustering based on similar activity patterns across time.