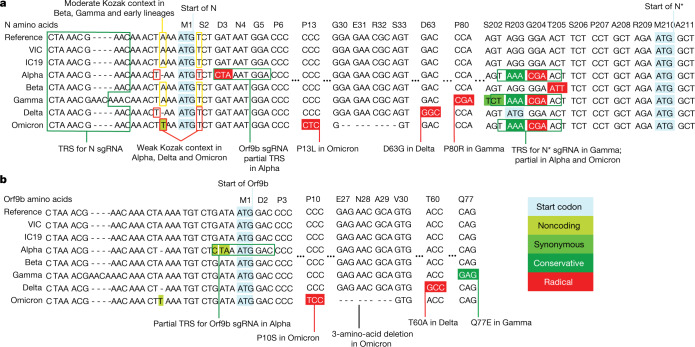
Fig. 6. VOCs present similar nucleotide mutations in N and Orf9b.
a, b, Genomic alignment of first-wave isolates and five VOCs showing sections of N and its 5′ region, codonized by CodAlignView in the reading frames of N (a) and Orf9b (b). The alignment includes TRS for N sgRNA present in all genomes; partial TRS for Orf9b sgRNA only in Alpha; TRS for N* sgRNA in Gamma and partial TRS in Alpha and Omicron. All mutations in Orf9b are colour-coded to indicate conservative (dark green) and radical (red) amino acid changes in Orf9b protein. We also highlighted a one-base deletion at 5′ of the N start codon in Alpha and Delta and an A to T substitution in Omicron, which change their adequate (A in −3, T in +4) Kozak initiation context to the weak (T in −3, T in +4) context, and could lead to more leaky scanning translation of Orf9b from the N sgRNA.