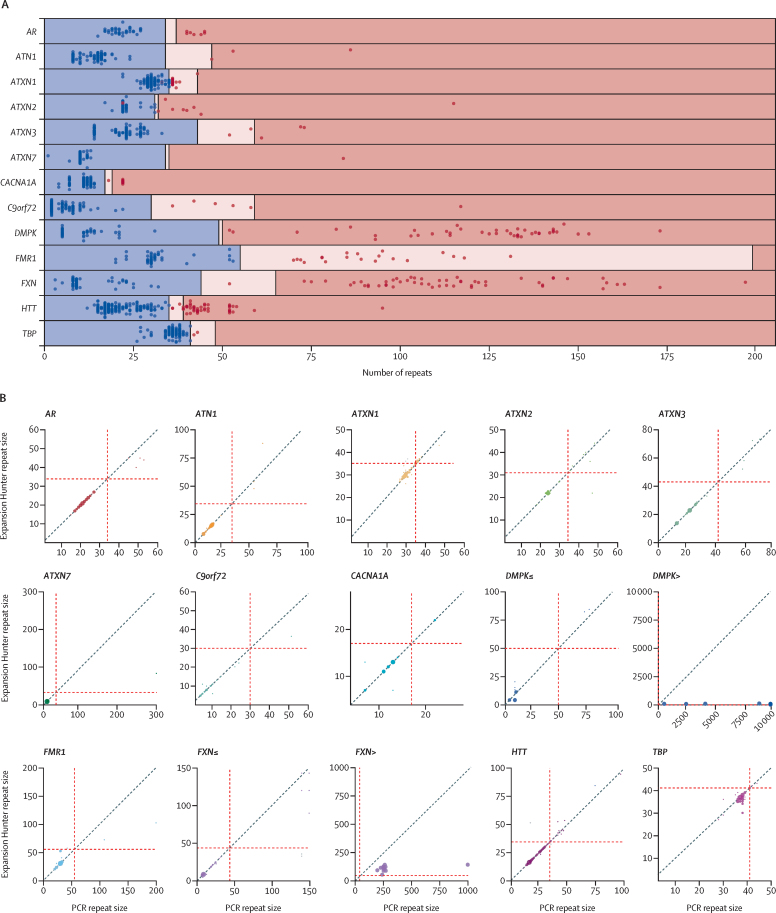
Figure 2.
Performance of repeat expansion detection using whole genome sequencing
(A) Swim lane plot showing sizes of repeat expansions predicted by ExpansionHunter across 793 expansion calls. Each genome is represented by two points, one corresponding to each allele for each locus, with the exception of those on the X chromosome (ie, FMR1 and AR) in males, for which only one point is shown. Points indicate the repeat length estimated by ExpansionHunter after visual inspection and the colours indicate the repeat size as assessed by PCR (blue represents non-expanded; red represents expanded). The regions are shaded to indicate non-expanded (blue), premutation (pink), and expanded (red) ranges for each gene, as indicated in the appendix (p 28). Blue points in pink or red shaded regions indicate false positives and red points in blue shaded regions indicate false negatives. The individual calls are provided in the appendix (p 27). (B) Repeat size correlation by locus. Bubble plots show PCR repeat sizes on the x axes and ExpansionHunter repeats sizes on y axes, with the size of each dot showing the number of patients with the same repeat size. The grey points visible for ATXN1, FMR1, FXN≤, and HTT represent ExpansionHunter estimations before visual inspection, whereas the corrected ExpansionHunter sizes after visual inspection are in colour. Red dashed lines represent the premutation cutoff for each locus (appendix p 28). FXN≤ and DMPK≤ show the repeat size correlation when the the size is less than or equal to the read length (ie, 150 bp). FXN> and DMPK> show the repeat size correlation when the size is larger than the read length.