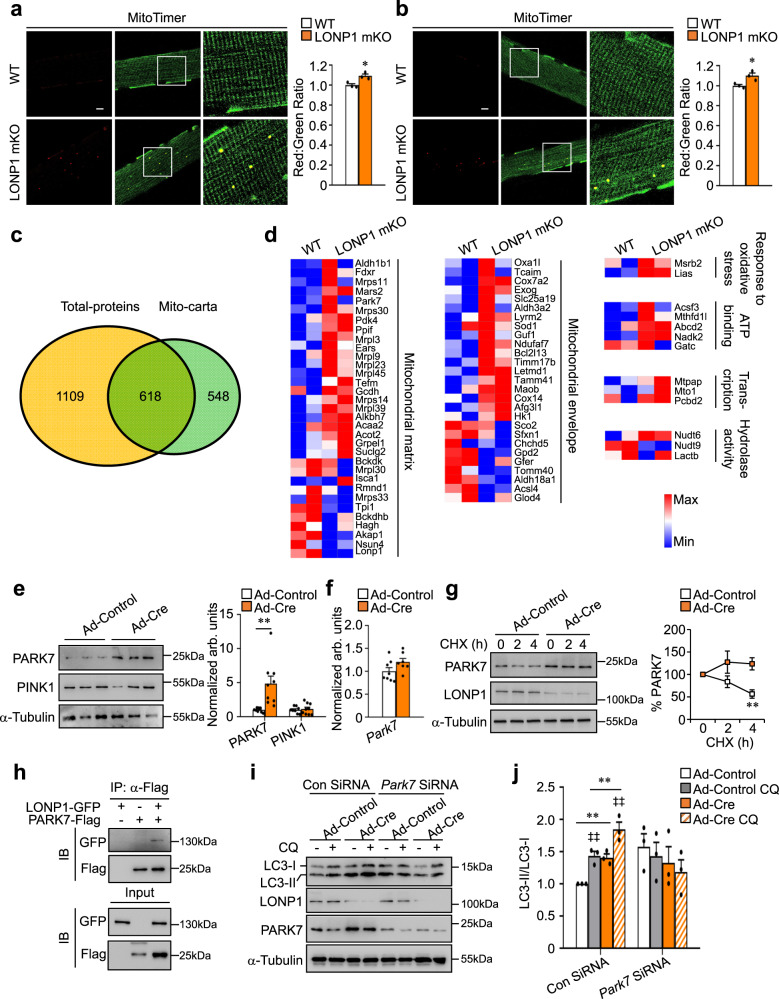
Fig. 6. LONP1 controls mitochondrial proteostasis that regulates muscle cell autophagy.
a, b (Left) Confocal images of TA (a) and EDL (b) muscles from LONP1 mKO/MitoTimer and Lonp1f/f/MitoTimer mice. The scale bar represents 10 μm. (Right) Quantification of MitoTimer Red:Green ratio. n = 3 mice per group. c, d Proteomes of crude mitochondria isolated from 2-week-old WT and LONP1 mKO muscles were analyzed by isobaric tags for relative and absolute quantification (iTRAQ) mass spectrometry. n = 2 independent samples per group. c Venn diagram representing the proteomics results compared with MitoCarta2.0 database. d Heat-maps analysis of mass spectrometry proteomics data. Individual proteins are shown to be regulated in the LONP1 mKO muscles as denoted by the color scheme. e (Left) Western blot analysis of myotubes subjected to adenovirus-based overexpression of Cre compared with control. (Right) Quantification of the PARK7/Tubulin and PINK1/Tubulin signal ratios. n = 3. f RT-qPCR analysis of Park7 in myotubes. n = 3. g (Left) Cycloheximide (CHX) treatment of myotubes for the indicated time. (Right) Quantification of the PARK7 protein levels is shown. n = 3. h Co-IP experiments were performed by cotransfecting LONP1-GFP and PARK7-Flag in HEK293 cells as indicated at the top. Antibody against the Flag epitope was used for co-IP. The extracts (Input) from the HEK293 cells and proteins from the IP were analyzed by immunoblotting (IB). Representative results for co-IP are shown. n = 3. i Western blot analysis of LC3-II/LC3-I ratios in myotubes transfected with control siRNA or Park7 siRNA and subjected to adenovirus-based overexpression of Cre compared with control. Myotubes were treated for 4 h with or without chloroquine (CQ) as indicated. j Quantification of the LC3-II/LC3-I signal ratios in (i). n = 3. Values represent mean ± SEM; for (a), (b), (e), (f) and (g), *P < 0.05, **P < 0.01 versus corresponding controls determined by two-tailed unpaired Student’s t test; for (j), **P < 0.01 versus corresponding controls, ‡‡P < 0.01 versus vehicle controls, determined by one-way ANOVA coupled to a Fisher’s LSD post-hoc test; Source data are provided as a Source Data file.