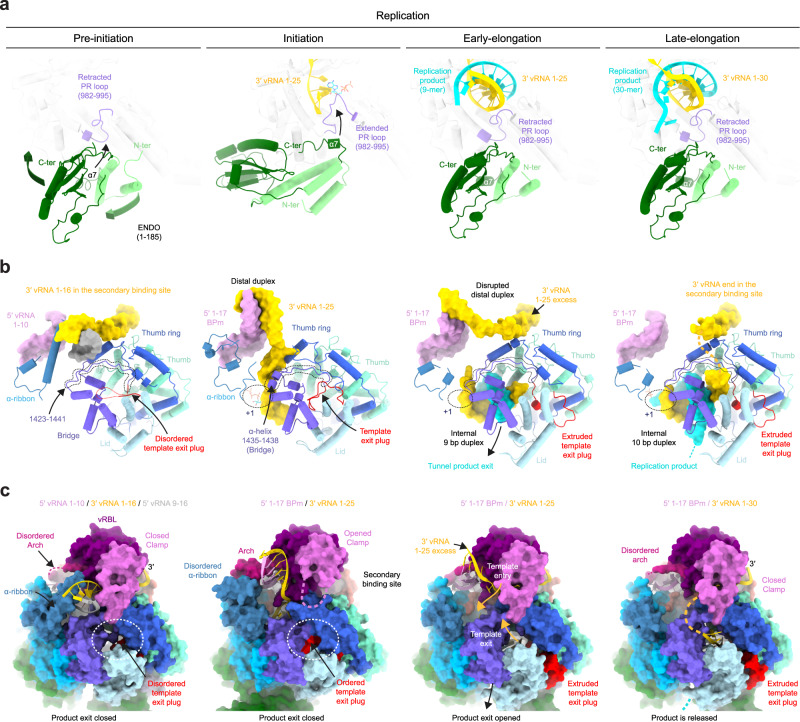
Fig. 3. Replication-induced conformational changes of LACV-LCItag_H34K.
a Endonuclease (ENDO) and prime-and-realign loop (PR loop) movements between the replication states. ENDO N-terminus (N-ter) and C-terminus (C-ter) are indicated. Arrows indicate movements of the ENDO, the α7 helix, and the PR loop. b LACV-L domain movements and RNA position variations between the replication states. For clarity, only domains that undergo conformational changes are displayed. The template exit plug, colored in red, is indicated with a dotted line if flexible. RNAs are shown as surfaces and colored as in Fig. 1c, d. Bridge residues 1423–1441 are surrounded by a dotted line. c Template exit plug, vRNA binding lobe (vRBL), clamp, and α-ribbon conformational changes between the replication states. LACV-L surface is displayed and each domain is colored as in Figs. 2a, b and 3b. The template exit tunnel is surrounded by a dotted line. Template entry/exit tunnels are indicated in the early-elongation state with gold arrows.