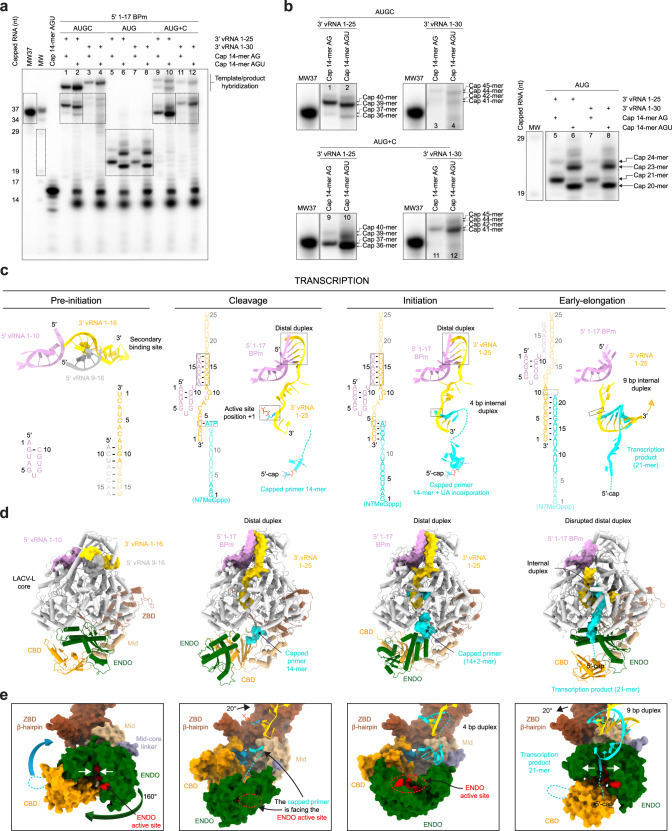
Fig. 4. Overview of LACV-LCItag_H34K activity and cryo-EM structures in transcription.
a In vitro transcription activity of LACV-LCItag_H34K using different combinations of 5′/3′-vRNAs, NTPs and capped primers (capped 14-mer finishing by “AG” (cap14AG) and in vitro produced capped 14-mer finishing by “AGU” (IVT cap14AGU)). The MW37 lane corresponds to a capped 37-mer identical in sequence to the theoretical transcription product of LACV-LCItag_H34K with the cap14AG and the 3′-vRNA1-25 without prime-and-realign. The lane indicated as MW corresponds to a RNAse T1 cleavage of MW37. Dotted squares indicate transcription products of interest that are reported in b. Source data are provided as a Source Data file. This experiment was repeated independently 3 times with similar results. b Zoom on LACV-LCItag_H34K transcription products. Lane numbers are referring to a. Transcription products length are indicated on the right side of the cropped gels with the corresponding molecular weight ladder. Source data are provided as a Source Data file. c Sequence and secondary structures of 5′/3′-vRNAs, capped RNA primer and transcription products. 5′-vRNAs, 3′-vRNAs and capped primer/product are respectively colored in pink/gray, gold and cyan. Bases present in the sequences but not seen in the structures are shown in transparent. d Cartoon representation of each LACV-LCItag_H34K transcription structure. Note that LACV-L in “capped primer active site entry” state has the same conformation as in “initiation” state. LACV-L domains are colored as in Fig. 1. RNAs are displayed as surfaces and colored as in c. e ENDO and CBD movements during transcription. The ENDO, CBD, Mid/ZBD, RNAs are displayed as surfaces and colored as in d. The ENDO active site and CBD cap-binding site are colored in red and blue. The closing/opening of product exit tunnel is highlighted by white arrows.