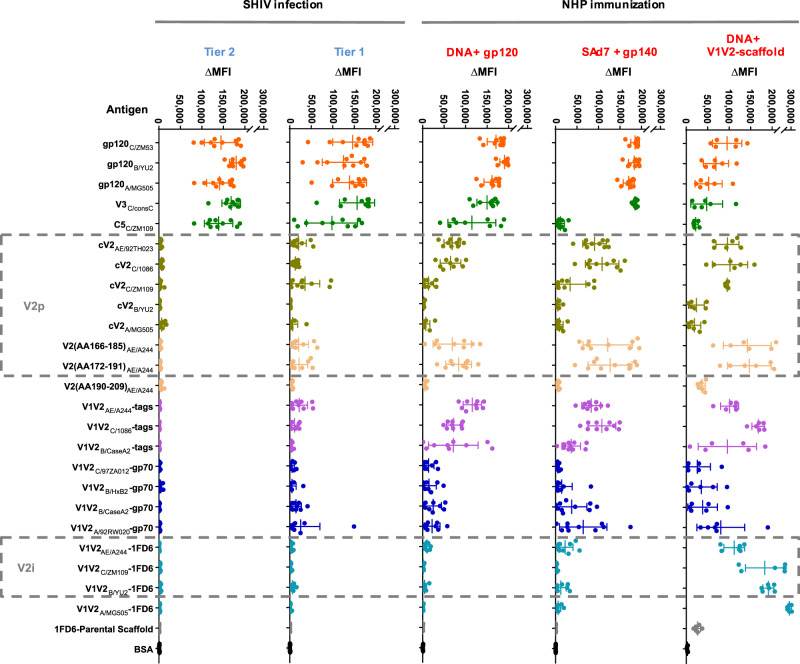
Fig. 3. Dot plot of Ab binding activities in plasma from infected and immunized NHPs assessed in the multiplex bead Ab binding assay.
Columns from left to right: Tier 2 SHIVC/1157-ipd3N4-infected NHPs (n = 10); Tier 1 SHIVC/1157ipEL-p-infected NHPs (n = 10); NHPs immunized with “DNA + gp120” (n = 9); NHPs immunized with “SAd7 + gp140” (n = 10); NHPs immunized with “DNA + V1V2-scaffolds” (n = 6). Each group was composed of biologically independent plasma specimens. Binding activity of plasma was measured against the same set of 24 Env antigens used in Fig. 1a; each row lists a tested antigen, and the rows are grouped by antigen type (peptide; gp120, etc.). Boxes with dashed lines denote antigens reactive with V2p- or V2i-specific mAbs. Beads identified as “1FD6 parental scaffold” (devoid of the V1V2 insert) and BSA (bovine serum albumin) were used as negative controls. Each dot represents the normalized mean fluorescence intensity (ΔMFI) generated with a plasma specimen diluted 1:200 from a single animal. Error bars indicate mean and standard deviation (SD). Colors of the dots represents a type of antigen (orange = gp120s, dark green = non-V1V2 linear peptides (V3 and C5), olive green = cV2 peptides, cream = linear V2 peptides, purple = V1V2-tags, dark blue = V1V2-gp70s, light blue = V1V2-1FD6s, gray/black = negative controls). Intensity of the reactivity is shown as the mean MFI calculated from experiments normalized on the basis of the reactivity of the mAb pool in each experiment from which background (PBS-TB) was subtracted. Data are represented as mean with error bars that indicate standard deviation (SD). Experiments were performed at least twice and in each experiment, samples were tested in duplicate. Source data are provided as a Source Data file. Abbreviations as in Fig. 1.