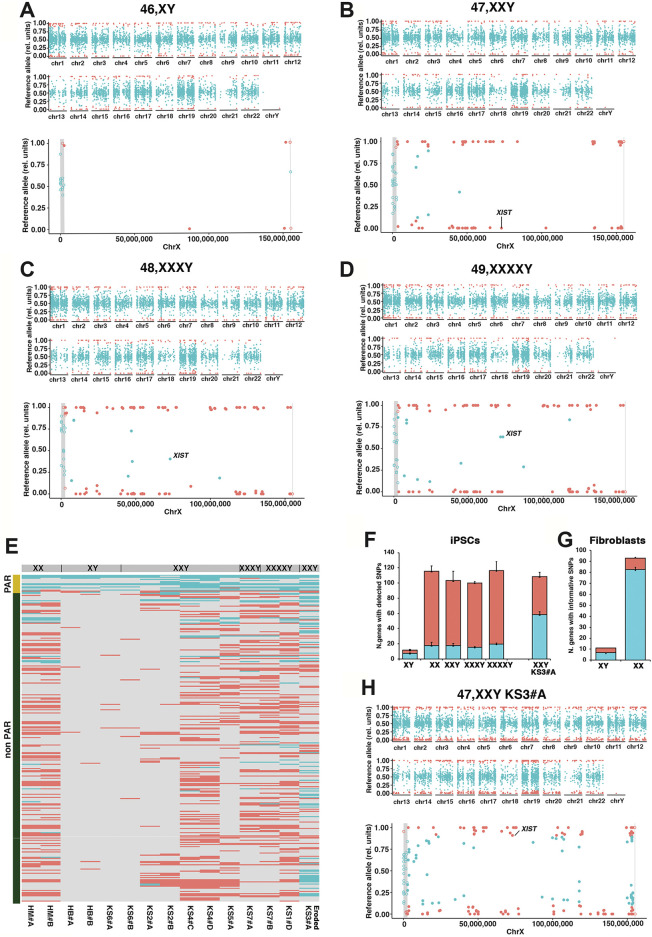
FIGURE 2.
Allele-specific expression (ASE) profiles of cases and controls iPSCs highlighting the tight retention of X chromosome inactivation. (A–D) Scatter plot profiles of coupled WES analysis and allele-specific RNA-Seq analysis performed on autosomal (upper panel) and X chromosomes (lower panel) showing the mono- (orange dots) or biallelic (light blue dots) gene expression status in four representative karyotypes. Gray rectangles indicate PAR1 and PAR2 regions, respectively. Solid dots indicate non-PAR genes; open dots show PAR genes. XIST allele-specific expression status is displayed for each sample. (E) Heatmap of mono- (orange) and biallelically expressed (light blue) genes in the PAR and non-PAR regions for the indicated iPSCs. The nonparametric Mann-Whitney tests showed the statistically significant differences between the active/inactive status of PAR versus non-PAR genes: p = 1.57e-12 for all genotypes, p < 2.22e-16 for the KS3#A eroded iPSCs. (F,G) Graphs showing the number of biallelic and monoallelic genes with at least one informative SNP for each genotype in iPSCs (F) and fibroblasts (G). Bars are medians ± standard deviations of two to three biological replicates for each sample. Two replicates of the 47,XXY KS3#A mildly eroded sample are shown. (H) Scatter plot profiles of the eroded 47, XXY KS3#A iPSCs sample.