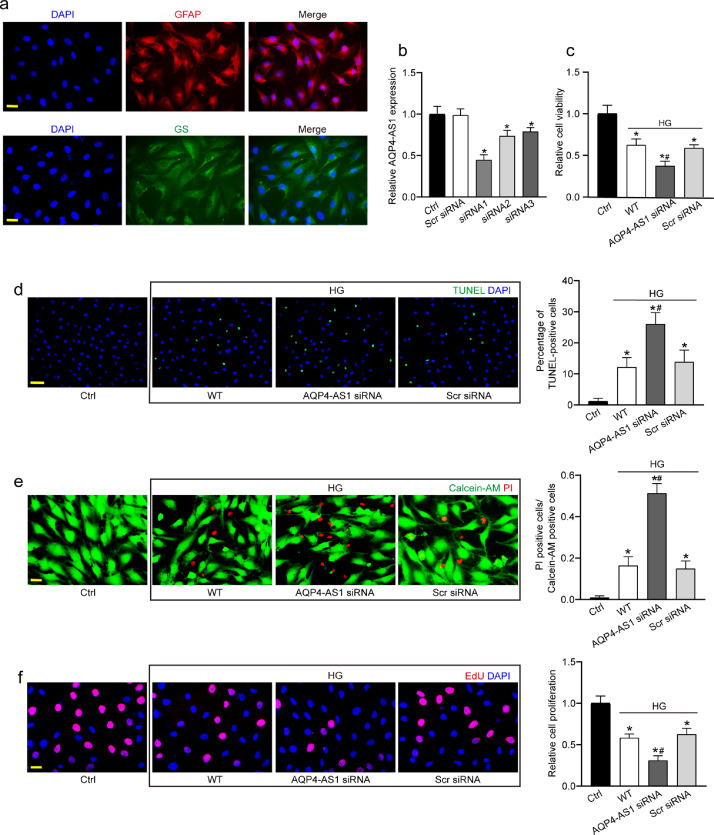
Fig. 2.
AQP4-AS1 regulates Müller cell function in vitro. (a) Primary Müller cells were identified with GFAP and GS protein immunofluorescence. GFAP, red; GS, green; DAPI, blue. Scale bar, 20 μm. (b) Müller cells were transfected with AQP4-AS1 siRNA1-3, scrambled (Scr) siRNA, or left untreated (Ctrl) for 48 h. qRT-PCRs were conducted to detect AQP4-AS1 expression (n = 4, *P < 0.05 versus Ctrl, one-way ANOVA followed by Bonferroni's post-hoc test). (c–f) Müller cells were transfected with AQP4-AS1 siRNA1, Scr siRNA, or left untreated, and then these cells were exposed with 30 mM high glucose for 48 h. The group without high glucose treatment was taken as the Ctrl group. The viability of Müller cells was determined by MTT assay (c, n = 4). Apoptotic cells were analyzed using TUNEL staining and quantitated.DAPI, blue; TUNEL, green. Scale bar 50 μm (d, n = 4). Apoptotic cells were also analyzed using Calcein-AM/PI staining, and quantitative analysis were conducted to detect the apoptotic percentage of Müller cells. Calcein-AM, green; PI, red. Scale bar, 20 μm (e, n = 4). EdU incorporation assay was performed to detect cell proliferation and the EdU positive cells were quantitated. DAPI, blue; EdU, red. Scale bar, 20 μm (f, n = 4). *P < 0.05 versus Ctrl; #P < 0.05 AQP4-AS1 siRNA versus Scr siRNA. The significant difference was determined by one-way ANOVA followed by Bonferroni test.