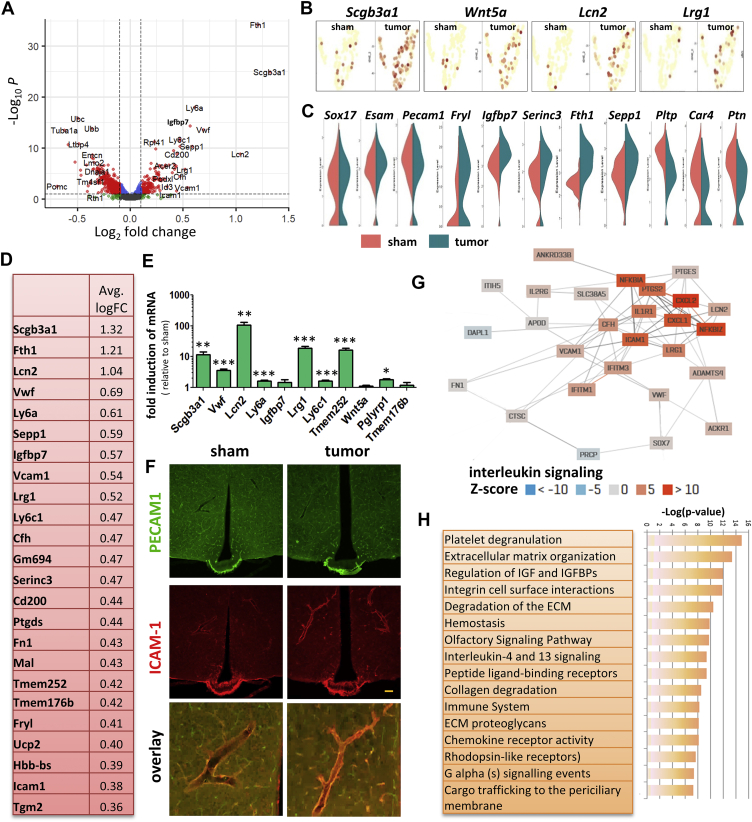
Figure 2.
MBH endothelial cell response to pancreatic cancer. Differentially expressed (DE) genes between sham and TB mice in endothelial cells as shown with a volcano plot (A). Featureplots showing the induction of genes in TB mice linked to the immune system (Lcn2, Lrg1), membrane organization (Scgb3a1), as well as secreted protein Wnt5a (B). Vln plots showing examples of highly upregulated DE genes, highly downregulated DE genes as well as conserved genes between sham and TB mice in the endothelial cluster (C). Top 24 upregulated genes in endothelial cells in TB mice sorted by fold inductions (D). qPCR analysis of top-induced genes using whole hypothalamic RNA extracts from sham and TB mice (E). IHC for ICAM-1 and PECAM1 in the MBH for sham and TB mice (F). PECAM1 labels all endothelial cells, while ICAM-1 is only expressed in a subset of the endothelial cells. Co-expression network using the top-induced genes in Lcn2+ endothelial cells revealing a network linked to inflammation. Z-scores indicate REACTOME pathway enrichment for interleukin signaling (G). Reactome pathway analysis of top 60 induced genes in endothelial cells of TB mice (H). qPCR values represent the mean ± SEM of at least three independent experiments and statistical significance between groups was determined with a Student's t-test (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). Scale bar = 100 μm.