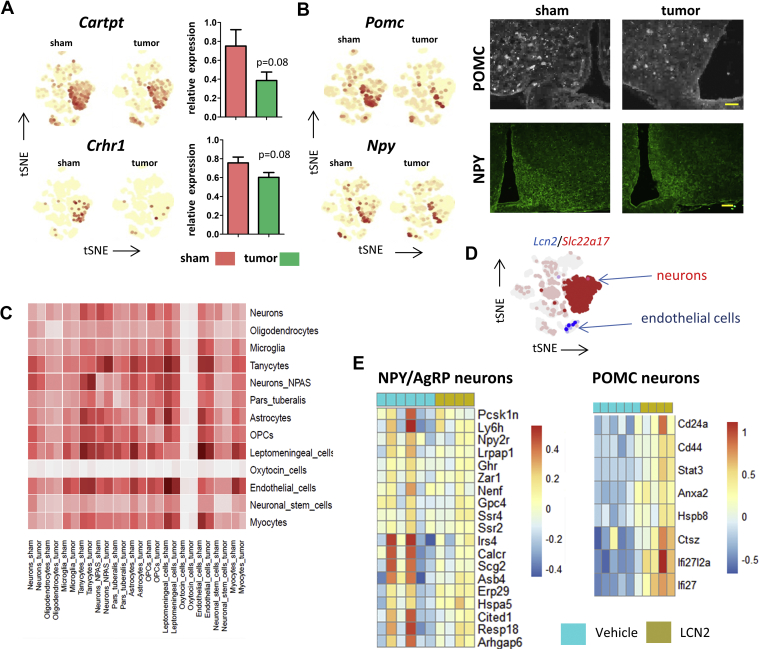
Figure 6.
Cell–cell communications in the MBH. Featureplots for Cartpt and Crhr1 for sham and TB mice as well as quantification by qPCR (A). Featureplots for anorexigenic Pomc and orexigenic Npy for sham and TB mice, as well as IHC for POMC and NPY in the MBH for sham and TB mice (B). Heatmap of the total number of ligand–receptor interactions between sham and TB mice. Each cell type is compared to cell types of the same condition (sham left, TB mice right) as found by scRNA-seq (C). Example of a potential cell–cell interaction in cancer cachexia with the receptor indicated in red and the ligand in blue (D). Heatmap for genes enriched in appetite stimulating NPY/AgRP neurons which are downregulated upon LCN2 treatment (left) and heatmap for genes enriched in appetite-decreasing POMC neurons which are upregulated after LCN2 treatment (right) (E). Enriched genes for NPY/AgRP neurons or POMC neurons were obtained by scRNA-seq using the top 100 enriched genes of the both subtypes of neurons as found by Campbell et al. 2017 [10]. qPCR values represent the mean ± SEM of at least three independent experiments and statistical significance between groups was determined with a Student's. Scale bar = 100 μm.