Significance
Here we address the important question of cross-talk between the mitochondria and cytosol. We show that the inner mitochondrial protein, MiNT, interacts with a protein on the outer mitochondrial membrane (mNT). This interaction occurs within the major outer membrane protein VDAC1. Inside the inner space of VDAC1, MiNT transfers its [2Fe-2S] clusters to mNT, which was shown to be a [2Fe-2S] cluster donor protein that donates its cluster(s) to apo-acceptor proteins residing in the cytosol. Hence, we suggest a pathway for transferring [2Fe-2S] clusters from inside the mitochondria to the cytosol.
Keywords: mitochondrial inner NEET protein (MiNT), CISD3, VDAC1, mitoNEET, [2Fe-2S] cluster
Abstract
Mitochondrial inner NEET (MiNT) and the outer mitochondrial membrane (OMM) mitoNEET (mNT) proteins belong to the NEET protein family. This family plays a key role in mitochondrial labile iron and reactive oxygen species (ROS) homeostasis. NEET proteins contain labile [2Fe-2S] clusters which can be transferred to apo-acceptor proteins. In eukaryotes, the biogenesis of [2Fe-2S] clusters occurs within the mitochondria by the iron–sulfur cluster (ISC) system; the clusters are then transferred to [2Fe-2S] proteins within the mitochondria or exported to cytosolic proteins and the cytosolic iron–sulfur cluster assembly (CIA) system. The last step of export of the [2Fe-2S] is not yet fully characterized. Here we show that MiNT interacts with voltage-dependent anion channel 1 (VDAC1), a major OMM protein that connects the intermembrane space with the cytosol and participates in regulating the levels of different ions including mitochondrial labile iron (mLI). We further show that VDAC1 is mediating the interaction between MiNT and mNT, in which MiNT transfers its [2Fe-2S] clusters from inside the mitochondria to mNT that is facing the cytosol. This MiNT–VDAC1–mNT interaction is shown both experimentally and by computational calculations. Additionally, we show that modifying MiNT expression in breast cancer cells affects the dynamics of mitochondrial structure and morphology, mitochondrial function, and breast cancer tumor growth. Our findings reveal a pathway for the transfer of [2Fe-2S] clusters, which are assembled inside the mitochondria, to the cytosol.
The iron–sulfur cluster (ISC) biogenesis system, located in eukaryotic cells inside the mitochondria, is where [2Fe-2S] ISCs are de novo synthesized by a complex system whose structure was elucidated in atomic resolution (1, 2). The [2Fe-2S] clusters produced by ISC are then transferred to different apo-acceptor proteins that either function as mitochondrial iron–sulfur proteins (3, 4), or as cluster donor proteins (1, 4, 5). The [2Fe-2S] clusters produced in the mitochondria need to reach cytosolic apo-acceptor proteins, including the cytosolic iron–sulfur protein assembly (CIA) pathway which is the biogenesis system responsible for [4Fe-4S] cluster formation (1, 5, 6). While ISC and CIA are well characterized (1, 7), the mechanism of export of [2Fe-2S] cluster from the mitochondria to the cytosol is still partially unknown (1, 8). Here we propose that two NEET [2Fe-2S] protein family members play a role in the transport of [2Fe-2F] clusters from the mitochondria to the cytosol.
The NEET family in humans is composed of three members: mitoNEET (mNT, CISD1), NAF-1 (CISD2), and the mitochondrial inner NEET (MiNT, CISD3) (9–11). The homodimeric mNT and NAF-1 proteins are located on the outer mitochondrial membrane (OMM); NAF-1 is also located at the endoplasmic reticulum (ER) membrane, and the ER–mitochondrial associated membranes (MAM) (10). MiNT is a soluble, monomeric protein, localized inside the mitochondria (12). The mNT and NAF-1 were recently shown to be important for mitochondrial function and morphology (13–15). They were also shown to play a role in cancer, diabetes, and Wolfram syndrome type 2 diseases (16). Downregulating any one of the NEET proteins, including MiNT, leads to increased accumulation of mitochondrial labile iron (mLI) and mitochondrial reactive oxygen species (mROS), as well as to disruptions in mitochondrial function and morphology (12, 14, 17). Mitochondrial iron and ROS accumulation were shown to affect mitochondrial respiratory capacity (12, 18–26). Since mitochondria are considered to play a central role in bioenergetics, metabolism, and other cellular pathways (27), mitochondria morphodynamics are tightly linked to the overall bioenergetic function of cells (28). The dynamics of mitochondrial architecture/morphology was shown to be involved in several cellular functions, including proliferation, viability, senescence, and signaling (28). The control of mitochondrial dynamics, through fusion or fission, displays therefore high plasticity in the regulation of mitochondrial hemostasis and metabolism. Mitochondrial morphology dynamical balance through fragmentation or elongation, in turn, depends on nutrient availability as well as ATP synthesis under the energy demands of the cell (28, 29).
Decreasing mNT or NAF-1 levels leads to a reduction of mitochondrial size indicative of a fission process in cells (24, 26). NAF-1 and mNT were previously shown to interact; in this interaction, mNT transfers its [2Fe-2S] cluster to NAF-1 (17). Moreover, both mNT and NAF-1 were shown to function as cluster donor proteins, transferring their [2Fe-2S] clusters to apo-acceptors in the cytosol, such as anamorsin, with NAF-1 showing higher transfer efficiency than mNT (30). The mNT was proposed to play an important role in regulating cytosolic iron regulatory protein 1 (IRP1) (31); mNT was also shown to bind within the central cavity of the voltage-dependent anion channel 1 (VDAC1), which suggested a potential role for mNT in regulating VDAC1 and maintaining the cellular iron/[Fe-S] hemostasis of cells (32). A major role of mitochondria in iron metabolism is thought to be iron–sulfur [Fe-S] cluster synthesis and transport to the cytosol (1, 33). Mitochondrial iron hemostasis is also considered a regulator of mitochondrial morphology (34, 35). However, the different players constituting the complete [2Fe-2S] transport machinery from the mitochondria to the cytosol have not been fully identified yet (7, 36, 37).
Here we show that the expression level of MiNT affects mitochondrial function and dynamics. Using human breast cancer cell lines with deceased or overexpressed levels of MiNT, we show that overexpressing MiNT is accompanied by mLI and mROS accumulation, as well as by heightened ROS resistance, cellular proliferation, and tumor growth. In contrast, suppression of MiNT expression results in mLI and mROS accumulation that are accompanied by cytosolic ROS accumulation and decrease in cellular proliferation and tumor growth. Using biochemical methods and computational studies, we further show that MiNT interacts with VDAC1, and, through VDAC1, with mNT, transferring its [2Fe-2S] clusters from inside the mitochondria to mNT that is facing the cytosol on the OMM. Our findings reveal the existence of a pathway for transferring of [2Fe-2S] clusters from within the mitochondria, to the cytosol.
Results
MiNT Overexpression in Human Breast Cancer Cells Enhanced ROS Resistance and Tumor Growth.
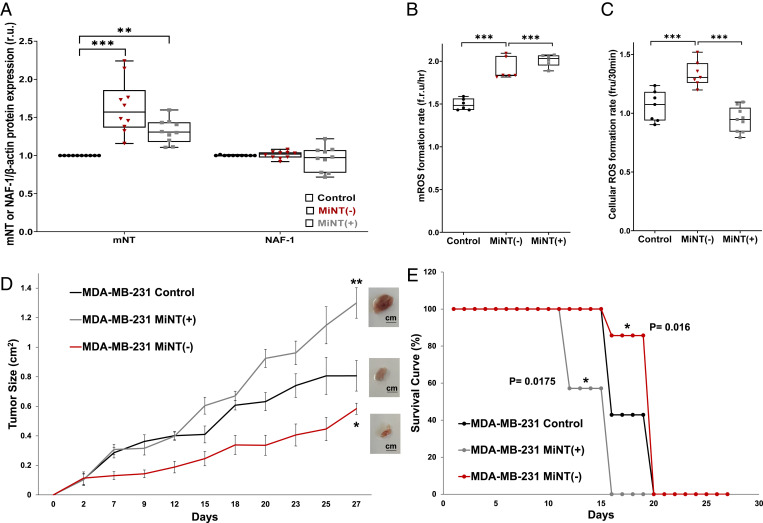
For our analyses of MiNT function, we chose triple-negative human breast cancer cells (MDA-MB-231). Breast cancer cells were chosen as they represent a major form of human epithelial cancers with a high demand for cellular iron (11). MDA-MB-231 cells were used to down-regulate MiNT protein (MiNT(−)) by ∼50%, or overexpress MiNT (MiNT(+)) by ∼10× (SI Appendix, Fig. S1A). As MiNT expression decreased by ∼50%, mNT expression increased by ∼60% (Fig. 1A). An increase in mNT expression also occurs when MiNT is overexpressed by ∼10×, yielding an mNT increase of ∼30%. On the other hand, NAF-1 level did not change in both MiNT cell lines compared to control cells (Fig. 1A).
Fig. 1.
The expression level of MiNT impacts the growth of human xenograft breast cancer tumors. (A) Enhanced expression of mNT protein but not NAF-1 in cancer cells with decreased—MiNT(−) (red dots)—or increased—MiNT(+) (gray dots)—MiNT protein levels; n = 7. (B) The mROS accumulation in MiNT(−) (red dots) and MiNT(+) (gray dots) cells; n = 7. (C) Cytosolic ROS accumulates in MiNT(−) (red dots) but not MiNT(+) (gray dots) cells; n = 7. Results from A–C are presented as box and whisker plots and include all data points measured in three different experiments. *P < 0.05, **P < 0.01, ***P < 0.001; Student’s t test. (D) Xenograft tumor growth (square centimeters) in mice injected subcutaneously with control MDA-MB-231 breast cancer cells (black), or MDA-MB-231 cells with high—MiNT(+) (gray)—or low—MiNT(−) (red)—MiNT expression. (Insets) Images of representative tumors; n = 7. *P < 0.05, **P < 0.01 (day 27). (E) Kaplan–Meier survival curve of mice injected with control MDA-MB-231 breast cancer cells (black), or MDA-MB-231 cells with high—MiNT(+) (gray)—or low—MiNT(−) (red)—MiNT expression; n = 7. Statistical significance was determined using log-rank (Mental–Cox) test to compare MiNT(−) or MiNT(+) to control; *P < 0.05.
Human breast cancer cells (MDA-MB-231) with decreased or increased expression of MiNT showed increased mROS accumulation (Fig. 1B), while ROS in the cytosol was increased only in the MiNT(−) line (Fig. 1C). This finding may result from enhanced ROS resistance of the MiNT(+) cell line, as shown in SI Appendix, Fig. S1B. As the cell biology characterization was conducted with human breast cancer cells, we elected to use a human xenograft model system to characterize the relationship between MiNT and tumor growth. Using the MiNT(−) and MiNT(+) MDA-MB-231 cell lines, xenograft breast cancer tumors were generated (Fig. 1D); MiNT(+) showed increased tumor growth compared to control, while MiNT(−) displayed decreased tumor growth compared to control or the MiNT(+) line (Fig. 1D). This finding correlated with enhanced survival of mice with MiNT(−) cells compared to control or mice with MiNT(+) cells, that showed the poorest survival (Fig. 1E). Interestingly, tumor growth rate of MiNT(−) tumors was affected by day 2, while that of control and the MiNT(+) tumors were similar until day 12. This finding could suggest that xenograft tumor establishment is dependent on MiNT function. Both lines (MiNT(−) and MiNT(+)) displayed high accumulation of mROS and mLI, that was corrected upon treatment of cells with the iron chelator deferiprone (DFP; SI Appendix, Fig. S1 C and D). DFP was also able to repair the impairment of both cells in mitochondrial membrane potential (MMP; SI Appendix, Fig. S1E). In future studies, we plan to study MiNT function in a murine tumor model system, as well as in three-dimensional human cultures.
MiNT Expression Modulates Mitochondrial Function and Dynamics through Stimulating Mitochondrial Fission or Fusion.
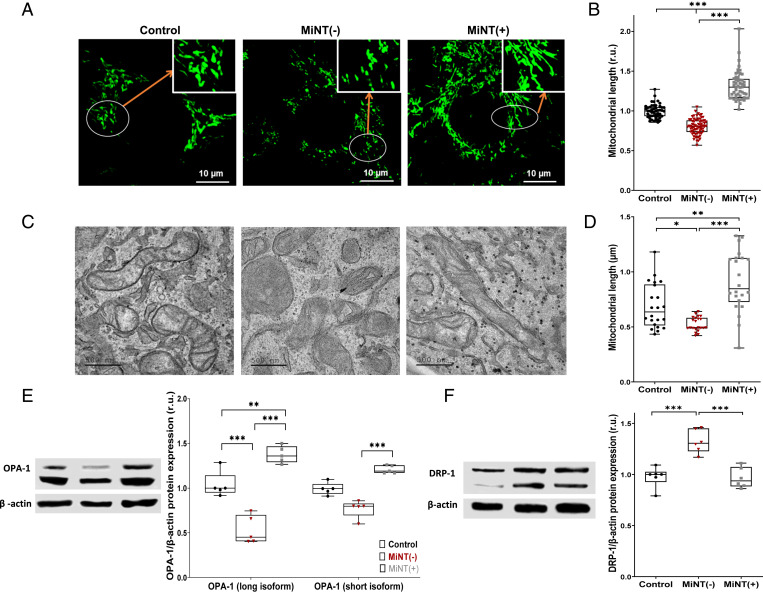
The expression level of MiNT was found to affect mitochondrial morphology and size, as well as the expression of OPA-1, a major protein involved in fusion, and DRP-1, a protein which is known to induce mitochondrial fission (38, 39). Fission of mitochondria was observed in MiNT(−) cells (Fig. 2 A–D), and this finding correlated with low expression levels of the OPA-1 protein, especially its large subunit (Fig. 2E), and high expression of DRP-1 (Fig. 2F). In MiNT(+) cells, mitochondrial fusion was observed (Fig. 2 A–D), and this finding correlated with high expression of OPA-1 (Fig. 2E) as well low expression of DRP-1 (Fig. 2F).
Fig. 2.
MiNT expression affects mitochondrial fusion and fission. (A) Representative confocal fluorescence microscope images of mitochondria from control, MiNT(−), and MiNT(+) cells. (B) Quantification of mitochondrial length in the different cell lines shown in A. (C) Representative TEM images of mitochondria from control, MiNT(−), and MiNT(+) cells. Scale bar is 500 nm. (D) Quantification of mitochondrial length in the different cell lines shown in A. Results in B and D are presented as box and whisker plots and include all data points measured in three different experiments. *P < 0.05, ***P < 0.001; Student’s t test. (E) Western blot analysis of OPA-1 levels in control, MiNT(−), and MiNT(+) lines. (Left) Representative blots and (Right) quantification graphs. β-actin was used for quantification. (F) Same as E but for DRP-1. Results in E and F are presented as box and whisker plots and include all data points measured in three different experiments. **P < 0.01, ***P < 0.001; Student’s T-test.
These changes in mitochondrial length were also associated with abnormal morphological structures of the organelle (SI Appendix, Fig. S2). In addition to these morphological and dynamical changes, functional changes were observed in mitochondria of the different cell lines. The decreased expression of MiNT was associated with high respiration and increased production of ATP, while overexpressing of MiNT did not alter respiration (SI Appendix, Fig. S3A). In contrast, glycolysis was increased in the MiNT(+) line and decreased in the MiNT(−) line (SI Appendix, Fig. S3B).
Taken together, our findings suggest that the expression level of MiNT impacts both mitochondrial dynamics and morphology by affecting mitochondrial function and expression level of DRP-1 and OPA-1.
MiNT Plays a Role in Transferring [2Fe-2S] Clusters from Inside the Mitochondria to apo-mNT Situated on the Mitochondrial Outer Membrane through the VDAC1 Channel.
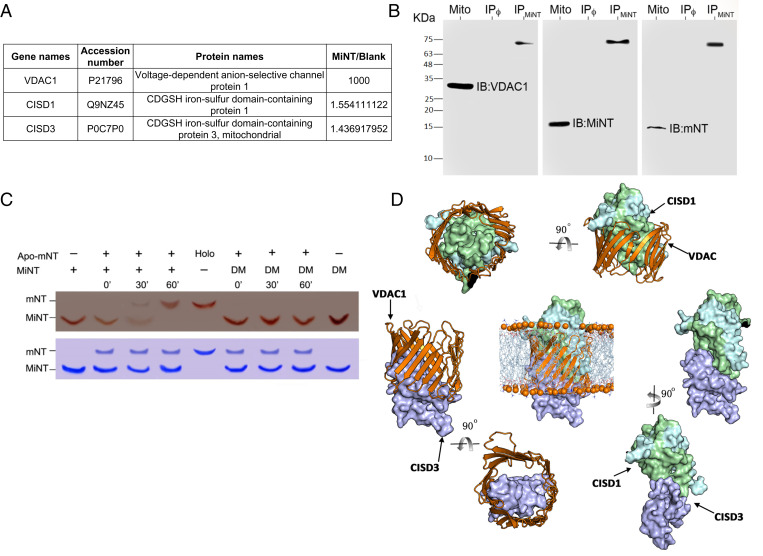
To identify mitochondrial MiNT-interacting protein partners, we used a coimmunoprecipitation (Co-IP) approach, with an antibody raised against the MiNT protein, applied to an enriched mitochondrial fraction obtained from MDA-MB-231 wild-type (WT) cells. The identification of the coimmunoprecipitated proteins was performed using mass spectrometry (MS) and Western blots. The MS analysis (SI Appendix, Table S1) revealed that MiNT interacts with VDAC1 and mNT (Fig. 3A). The Co-IP of VDAC1, MiNT, and mNT was then verified by Western blot analysis using antibodies raised against MiNT, mNT, and VDAC1 (Fig. 3B). Furthermore, there was specific interaction with each antibody on the control mitochondrial fraction (Mito), and the three interacting proteins revealed the same band at a higher molecular weight (IPMiNT), corresponding to the weight of the three-protein complex (Fig. 3B). NEET proteins are known to bind [2Fe-2S] clusters, and, due to the high homology of NEET proteins and localization of MiNT, we hypothesized that MiNT is also able to donate its [2Fe-2S] clusters to mNT. Using native gels and purified NEET proteins, an in vitro [2Fe-2S] cluster transfer assay was performed between MiNT and apo-mNT. This assay (Fig. 3C) showed that MiNT transfers its [2Fe-2S] clusters to apo-mNT (Fig. 3C). Taken together with our recent report that mNT interacts with VDAC (32), our findings shown in Fig. 3 demonstrate that a complex of MiNT, VDAC, and mNT can form and that this complex can facilitate cluster transfer between MiNT and mNT.
Fig. 3.
MiNT interacts with VDAC and mNT and transfers its [2Fe-2S] cluster to apo-mNT. (A) Identification of VDAC1, mNT, and MiNT proteins following Co-IP with an anti-MiNT antibody applied to an enriched mitochondrial fraction from MDA-MB-231 cells (SI Appendix, Table S1). (B) Western blots of VDAC1/MiNT/mNT after Co-IP of mitochondrial enriched fraction from MDA-MB-231 WT cells, using MiNT, VDAC1, or mNT antibodies. The presence of VDAC1, MiNT, or mNT was detected on the mitochondrial protein extract (Mito), or on protein G beads that were preincubated with MiNT antibody (IPMiNT) or not (IPϕ). (Left) Blot treated with anti-VDAC1 alone, showing the VDAC1 protein band at the molecular weight 30 KDa in the Mito line, (Middle) blot treated with anti-MiNT antibody showing the band in the Mito line at 14 KDa, and (Right) blot treated with anti-mNT antibody showing a specific binding at the Mito line at 12 KDa. All three membranes showed a higher molecular weight band at the same size in the IPMiNT line of about 60 KDa, which indicated the presence of the VDAC1–MiNT–mNT complex. (C) (Upper) Native gel showing the holo-NEET protein as a red band due to the presence of [2Fe-2S] cluster imbedded in the protein. The mNT protein is the upper band, while MiNT protein is the lower band. Holo-MiNT is shown to donate its clusters to apo-mNT after 30 min of incubation with apo-mNT in the presence of a reducing agent. DM is the double-mutated form of MiNT that is very stable and is not able to donate its [2Fe-2S] clusters; it is used as a negative control for the [2Fe-2S] transfer between the two NEET proteins. (Lower) A duplicate gel stained with Coomassie for protein levels. (D) The computationally constructed ternary binding complexes; mNT binds with VDAC1 mainly via the identified site 2 and part of site 1, and uses its large area of site 1 to interact with site1 of MiNT. Besides, site 2 and site 3 of MiNT are the major areas that contribute to the contacts with VADC1.
Computational Modeling of MiNT–VDAC–mNT Interaction.
To characterize how MiNT, VDAC, and mNT interact, a computational prediction and modeling framework was developed to define this ternary complex and inspect its recognition process (see the details in SI Appendix and Movie S1). First, we used our in-house protein–protein interaction (PPI) binding site identification method Fd-DCA (40, 41) to search potential sites on the surface of the soluble domain of mNT (homodimer) and MiNT. Fd-DCA is a method for PPI binding site identification combining fragment docking-based searching and direct-coupling analysis. MiNT and mNT belong to the same protein family; hence, distinguishing the interprotein residue–residue coevolutionary couplings from intraprotein signals is challenging. In this work, we only used the fragment docking-based searching module of Fd-DCA to identify the candidate binding sites. We selected a group of residues to locate the candidate binding sites for the next step of protein–protein docking simulations. As a result, we obtained three candidate binding sites for each of them, as shown in SI Appendix, Fig. S4. The potential site 1 and site 2 of mNT are almost symmetric, having significant overlapped residues. The potential site 3, located on the bottom of the protein away from the ISC, was not considered as a potential interacting site in this work. The residue composition of each site is shown below each structure in SI Appendix, Fig. S4. Then we used high-ambiguity-driven protein-protein docking (HADDOCK) (42), a protein–protein docking webserver, to predict the binding poses for the PPI pairs for VDAC1, mNT, and MiNT. For each candidate site of mNT and MiNT, we chose two residues located around the center position of the candidate site as the constraints (representatives for the location) of the active residues required as the constraints for the docking simulations. For VDAC1, we did not specify the active sites or residues, we randomly picked the residues scattered on the inner surface of the tunnel, because any protein which interacts with VDAC1 should insert into its large tunnel, forming a circle of contacts with the inner residues of VDAC1. Hence, any potential sites for VDAC1 were not used as constraints. We constructed a ternary complex of VDAC1–mNT–MiNT by selecting representative conformations from the largest clusters of docked binary conformations, and refined its binding conformation by using the steepest descent energy minimization approach. The constructed ternary binding complexes are shown in Fig. 3D. The mNT binds with VDAC1 mainly via its site 2 and part of site 1, and uses its large area of site 1 to interact with site 1 of MiNT. Site 2 and site 3 of MiNT are the major areas potentially interacting with VADC1. Finally, we used a coarse-grained molecular dynamics simulation to describe the recognition process between mNT and MiNT via VADC1 (Movie S1 and SI Appendix, Fig. S5). This movie shows that the relatively smaller protein MiNT can quickly tunnel into one side of VDAC1 and form a stabilized binary complex, then mNT finds a suitable orientation and inserts itself into the VDAC1 from the other side of VDAC1 until it forms a well-packed ternary complex (Movie S1).
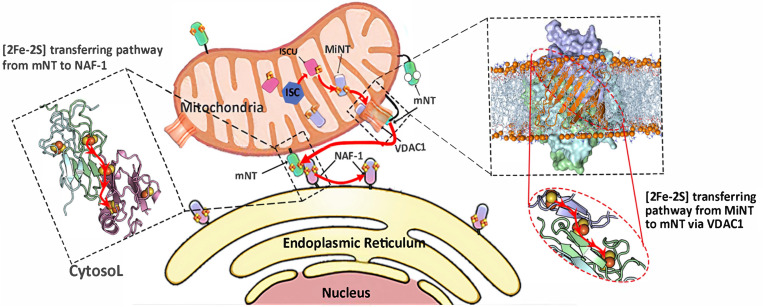
Summarizing our biochemical and computational studies, we propose a model for a chain of [2Fe-2S] transfer from inside the mitochondria from holo-MiNT to apo-mNT through the VDAC1 channel, then from holo-mNT to apo-NAF-1 in the MAM area, or from holo-mNT or holo-NAF-1 to any other apo-accepter in the cytosol (Fig. 4).
Fig. 4.
Proposed model for the VDAC1-mediated NEET protein chain transfer of [2Fe-2S] between the mitochondrial and the cytosol. (Right) The proposed chain that presents the interaction between MiNT, mNT, and VDAC1. (Middle) The transfer of the cluster from holo-mNT to apo-NAF-1. (Left) The transfer of the [2Fe-2S] cluster from the mNT and NAF-1 to apo-accepters in the cytosol.
Discussion
MiNT is the last human NEET protein member to be characterized. Its function was shown to be associated with mitochondrial iron/[Fe-S] hemostasis (12, 14), cancer proliferation, and ferroptosis (14). MiNT was also found to have a role in apoptosis (43). Altering the expression of MiNT affects mROS as well as cytosolic ROS. MiNT(−) cells accumulate high ROS inside and outside of their mitochondria which is associated with decreased size of xenograft tumors. In contrast, MiNT(+) had high growth of xenograft tumors, potentially due to MiNT(+) cells having higher ROS resistance and higher expression levels of mNT that is known to protect cells and mitochondria from oxidative stress (44, 45). Previously, mNT and NAF-1 were shown to play a role in mitochondrial morphology and dynamics (13, 16, 24, 27). Here we show that MiNT is also involved in mitochondrial function, morphology, and dynamics. In MiNT(−) cells, fission was associated with low expression of the OPA-1 fusion-inducing protein, and high expression of DRP-1, a fission-promoting protein. Conversely, high expression of MiNT was associated with higher fusion of mitochondria and increased expression of OPA-1. These features suggest a strong role for MiNT in regulating the mitochondrial fission/fusion pathway.
NEET proteins harbor a labile [2Fe-2S] cluster that can be transferred to many different apo-acceptor proteins. The mNT and NAF-1 were shown to interact and contribute to the same pathway, through the ability of mNT present at the OMM to donate its cluster to NAF-1 that is mainly present on the ER (17). In addition, mNT was found to interact with VDAC1 and play a role in [Fe-S]/iron hemostasis between the mitochondria and the cytosol (32). MiNT (CISD3) is the only member of the human NEET family to localize within the mitochondria where the cellular biosynthesis of [2Fe-2S] clusters takes place. Many studies focused on the export of [2Fe-2S] clusters from the mitochondria to the cytosol, but, to date, the exact mechanism for this process remains incomplete. Here we show that MiNT is able to transfer its [2Fe-2S] clusters to mNT when both proteins interact inside the VDAC1 channel. Co-IP with anti-MiNT antibodies, MS, and computational modeling of complex formation bring significant support to this pathway. Our work highlights a possible route for [2Fe-2S] clusters to be transferred from the mitochondria to the cytosol—via MiNT to mNT cluster transfer inside the VDAC1 protein (Fig. 4). It further supports the involvement of NEET proteins in [2Fe-2S] cross-talk between the mitochondria and cytosol. The existence of this pathway and the central role it plays in cellular metabolism, regulating iron, ROS, and overall Fe-S metabolism can affect our understanding of many different biochemical, molecular, and cellular processes key to diseases such as diabetes, cancer, and neurodegeneration.
Materials and Methods
Cell Growth.
MDA-MB-231 human breast cancer cells were obtained from ATCC and propagated as previously described (18). Plasmids containing short hairpin RNA (shRNA) in pGFP-RS vector (OriGene Technologies, Inc.) were used for repressing MiNT expression (−), whereas plasmids containing the pEGFP-N1 vector (Clontech Laboratories, Inc.) were used for overexpressing MiNT(+). Cell growth and transfection were performed as described earlier (12, 18, 22).
Animal Experiments.
Animal experiments were performed in compliance with the Hebrew University Authority for biological and biomedical models (NS-17-15262-4). Xenograft tumor of MDA-MB-231 human breast cancer cells were generated after injecting (2.5 × 106) into female athymic nude (FOXN1NU) mice of 5- to 6-wk age. Cells injected included normal Control, shRNA MiNT [MiNT(−)], and overexpression MiNT [MiNT(+)] MDA-MB-231 lines. Cells were injected subcutaneously to the flack of each mouse. Mice weight and tumor size were measured twice a week as described previously in ref. 22. All results were presented as mean ± SEM.
Protein Blots.
For protein blot analyses, cells were grown to full confluence, washed twice with 1× phosphate-buffered saline (PBS) and immediately scraped off the plate into a microcentrifuge tube with 1× Laemmli sample buffer and heated to 95 °C for 10 min. Protein gels were loaded with equal amounts of proteins and analyzed using antibodies against NAF-1, mNT, MiNT (18, 21), β-actin (R&D Systems, MAB8929), DRP-1 (Abcam, ab56788), and OPA-1 (Abcam, ab157457). For DRP-1 and OPA-1 antibodies, we used radioimmunoprecipitation assay (Sigma-Aldrich) buffer for lysing the cells, then lysates were kept on ice for 2 h with vortex every 30 min, then centrifugation at 15,000 rpm for 15 min, and supernatant was used for protein denaturation with Laemmli sample buffer 5×. Protein gels were loaded with equal amounts of protein, determined using The Pierce 660 nm Protein Assay (Thermo Scientific, cat. no. 22662) (46); Goat Anti-Rabbit IgG, H & L Chain Specific Peroxidase Conjugate (Sigma, 401315), and Peroxidase-conjugated AffiniPure Goat anti-mouse IgG (H+L) (Jackson ImmunoResearch Laboratories, AB_10015289) were used as secondary antibodies (7, 23). All experiments were repeated at least three times.
Fluorescence Microscope.
Cells were cultured and imaged by epifluorescent microscopy; for MMP, we used TMRE (tetramethylrhodamine ethyl ester) (Sigma, 87917) at a concentration of 0.1 μM. The mLI was determined with the fluorescent probe RPA (rhodamine B-[(1,10-phenanthrolin-5-yl) aminocarbonyl] benzyl ester) (Squarix Biotechnology, ME043.1) as described in ref. 18. The mROS accumulation was determined using mitoSOX Red (Invitrogen, M36008) according to refs. 22 and 46. Images were analyzed with Volocity (Quorum Technologies Inc.) and/or with Image-J (18). For cell ROS measurements, CellROX Deep Red Reagent detection assay (Invitrogen, C10422) was used in conjunction with a Biotek plate reader (excitation 640 nm, emission 665 nm). Quantification of mLI was performed using 30 different fields (five cells per field). Quantification of mitoSOX and cellular ROS, and fluorescence changes was performed by analyzing nine different fields (five cells per field). The mLI and mROS levels were averaged from three independent experiments. Cells were plated onto microscope slides glued to perforated 3-cm-diameter tissue culture plates as previously described (18, 46). Cells were also incubated with or without DFP (3-Hydroxy-1,2-dimethyl-4(1H)-pyridone; Sigma, 379409) at a concentration of 100 μM for 1 h prior to measuring for MMP, mLI, and mROS as described in refs. 18 and 21. For the ROS resistance experiment, we used DHR-123 (Dihydrorhodamine) probe (Biotium, cat. no. 10055) concentration 50 µM for 15 min at 37 °C, washed 2×, then read the signal after stabilizing for 15 min, then added H2O2 in a concentration of 50 µM, and continued the ROS increasing for 60 min (22). For mitochondrial length fluorescence imaging, cells were treated with the mitochondrial tracker Rhodamine 800 (Sigma, 83701). Cells were then evaluated by the Olympus FV3000 confocal laser-scanning microscope, and all images were analyzed with ImageJ.
Mitochondrial Bioenergetics, Oxygen Consumption Rate, Cellular Glycolysis, and Extracellular Acidification Rate.
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse XFp, Agilent, cell mito-stress analyzer (Agilent Technologies, Inc.) with the XF Cell Mito Stress Kit and XF Glycolysis Stress Kit (Agilent Technologies, Inc.), as previously described (26), and according to the manufacturer’s instructions (40). MDA-MB-231 cells (25,000) were grown to ∼80% confluence in complete medium overnight before the experiment. The initial medium was then exchanged with a seahorse-running medium consisting of Dulbecco’s modified Eagle’s medium base without glucose, L-glutamine 2 mM, sodium pyruvate 1 mM, glucose added to a final concentration of 25 mM, and pH adjusted to be 7.3 to 7.4. Then microplates containing cells were incubated at 37 °C without CO2 for 1 h before the assay. Plates were then placed into the XFp analyzer. The OCR was calculated after the sequential additions of oligomycin A 1.5 µM, FCCP 0.75 µM, antimycin A/Rotenone 1 µM, using an XF Cell Mito Stress Test kit. The ECAR medium initially did not contain glucose, and then was measured after the addition of glucose 10 mM, oligomycin A 2 µM, and 2-DG 50 mM. Results were expressed as mean ± SD of three independent experiments. All measurements were recorded at set interval time points. All materials and compounds were obtained from Seahorse Bioscience (18). Calculations of the OCR and the ECAR tests parameters were performed according to the manufacturer recommendations, using the equations described in ref. 47.
Transmission Electron Microscopy for Mitochondrial Structure.
Cells were grown on eight-well Permanox chamber slides and, upon reaching ∼80% confluence, were fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.4) for 2 h at room temperature, then prepared for transmission electron microscope (TEM) imaging as described in detail in refs. 18 and 26. Results were evaluated from 5 to 10 different cells randomly selected, averaged over 10 to 20 fields per cell; at least 100 to 200 mitochondria were counted. Mitochondrial damages were expressed as the ratio of damaged organelles to total number of organelles, in three independent experiments (16, 40). Mitochondrial length was similarly determined in three different biological repeats (16, 40).
Mitochondrial Fraction Enrichment and Protein Solubilization.
Mitochondria were isolated following the protocol published by Lampl et al. (48). Briefly, 3.0 × 106 cells were collected and washed with precooled PBS 1× twice and centrifuged at 600 × g for 5 min at 4 °C. The cell pellet was resuspended into 1 mL of mitochondrial isolation buffer (250 mM Mannitol, 70 mM sucrose, 10 mM Tris/HCl, pH 7.4, and 0.1 mM [ethylenebis(oxyethylenenitrilo)]tetraacetic acid/Tris) and homogenate using a glass dounce homogenizer. Then the cells were broken using a syringe and a 27-gauge 1/2-inch needle. The cell debris were centrifuge at 400 × g, and the supernatant containing the mitochondria crude was collected. The mitochondria crude extra was centrifugated at 10,000 × g for 10 min at 4 °C, and the pellet containing the enriched mitochondrial fraction was resuspended in mitochondrial isolation buffer at 4 °C. Enriched mitochondrial fraction was then solubilized in 50 mM Tris-pH-7.4, 150 mM NaCl, 1% Nonidet P-40, protease inhibitors mixture (P8340) 1:100, phosphatase inhibitors 1:100, and 1 mM NaV04 1:200. The lysate was then centrifuge at 5,000 × g for 10 min, the supernatant containing the solubilized protein was recovered, and the protein concentration was measured.
Immunoprecipitation.
Protein G beads were incubated with polyclonal anti-MiNT antibody (4 µg/mL) in 50 mM Tris-pH-7.4, 150 mM NaCl, 1% Nonidet P-40, protease inhibitors mixture (P8340) 1:100, phosphatase inhibitors 1:100, and 1 mM NaV04 1:200. After five washings in the same buffer, 400 µg to 500 µg of protein were incubated with the conjugated beads for 3 h to 4 h at 4 °C. The beads were then washed three times with the same buffer (without inhibitor) and two times in 50 mM Tris and 150 mM NaCl. After final pelleting at 5,000 × g for 5 min, the beads were analyzed by MS or by Western blotting as described above and below.
Sample Preparation for MS Analysis.
After the last step of immunoprecipitation, beads were washed twice with 150 mM NaCl, 50 mM Tris⋅HCl pH 7.4. Packed beads were then resuspended in 100 μL of 8M urea, 10 mM dithiothreitol (DTT), 25 mM Tris⋅HCl pH 8.0 and incubated for 30 min at 22 °C. Next, iodoacetamide (55 mM) was added, and beads were incubated for 30 min (22 °C, in the dark), followed by addition of DTT (20 mM). The urea was diluted by the addition of six volumes of 25 mM Tris⋅HCl pH 8.0. Trypsin was added (0.3 μg per sample), and the beads were incubated overnight at 37 °C with gentle agitation. The beads were spun down, and the peptides in the supernatant were desalted on C18 homemade Stage tips.
nanoLC-MS/MS Analysis.
MS analysis was performed using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) coupled online to a nanoflow ultra high performance liquid chromatography instrument, Ultimate 3000 Dionex (Thermo Fisher Scientific). Peptides (0.35 μg, as estimated by optical density 280 nm) were separated over a nonlinear 60-min gradient (0 to 80% acetonitrile) run at a flow rate of 0.3 μL/min on a reverse-phase 25-cm-long C18 column (75 μm ID, 2 μm, 100 Å, Thermo PepMapRSLC). The survey scans (380 to 2,000 m/z, target value 3E6 charges, maximum ion injection time 50 ms) were acquired and followed by higher-energy collisional dissociation–based fragmentation (normalized collision energy 25). A resolution of 70,000 was used for survey scans, and up to 15 dynamically chosen most abundant precursor ions, with “peptide preferable” profile, were fragmented (isolation window 1.6 m/z). The MS/MS scans were acquired at a resolution of 17,500 (target value 1E5 charges, maximum ion injection time 120 ms). Dynamic exclusion was 60 s. Data were acquired using Xcalibur software (Thermo Scientific). To avoid a carryover, the column was washed with 80% acetonitrile, 0.1% formic acid for 25 min between samples.
MS Data Analysis.
Mass spectra data were processed using the MaxQuant computational platform, version 1.5.3.1254. Peak lists were searched against translated coding sequences of the human proteome obtained from Uniprot. The search included cysteine carbamidomethylation as a fixed modification and oxidation of methionine as variable modifications, and allowed up to two miscleavages. The match-between-runs option was used. Peptides with a length of at least seven amino acids were considered, and the required false discovery rate was set to 1% at the peptide and protein level. Protein identification required at least two unique or razor peptides per protein. Relative protein quantification in MaxQuant was performed using the label-free quantification algorithm. Protein contaminants and proteins identified by less than two peptides were excluded from the analysis.
Protein Purification and Preparation of apo-mNT.
The mNT and MiNT proteins were expressed and purified as described in refs. 12 and 49–58. Briefly, the soluble part of mNT protein (residues 33 to 108) and MiNT (residues 36 to 127) were expressed in Escherichia coli BL21-RIL grown in lysogeny broth supplemented with 30 μg/mL kanamycin and 34 μg/mL chloramphenicol. At an O.D. 600 nm of 0.6, the cells were supplemented with 0.75 mM FeCl3 (only for mNT protein), and the expression was activated using 0.25 mM of isopropyl ß-D-1-thiogalactopyranoside. Cell growth proceeded for an additional 12 h at 310 K. From lysed cells, the mNT proteins were purified using Ni-agarose and size exclusion chromatography as described in ref. 49. MiNT protein was purified by two consecutive rounds of ion exchange chromatography followed by size exclusion chromatography as describe in ref. 12. Apo-mNT was obtained by incubating the purified protein in 20 mM Bis-Tris pH 6.0, 100 mM NaCl until the protein became colorless, and the protein was then dialyzed against 20 mM Tris pH 8.0 and 100 mM NaCl.
Native Gel [2Fe-2S] Cluster Transfer Assay.
The [2Fe–2S] cluster transfer was analyzed by absorption spectroscopy and native polyacrylamide gel electrophoresis, as described in ref. 17, with a native gel with 3% acrylamide.
Statistics.
Statistical significance tests (Student’s t test) for protein expression and analysis of epifluorescent microscope and TEM images were performed using GraphPad Prism 8.3.1 software. Results are presented as mean ± SE including all measured data points, or as box and whiskers plots including all measured data points; the line inside the box represents the median, the box represents the interquartile range, and the whiskers represent the range. Differences were statistically significant if the Student’s t test produced a probability value of less than 5% (*P < 0.05; **P < 0.01; ***P < 0.001).
Computational Prediction of Binding Mechanism among mNT–VDAC–MiNT.
To understand how these three proteins interact with each other and help us understand the [2Fe-2S] cluster transferring pathway, we firstly combined protein–protein binding site identification method Fd-DCA with the protein–protein docking tool HADDOCK to predict the binary complexes of mNT–MiNT, mNT–VDAC1, and MiNT–VDAC1, and build the ternary complex of VDAC1–MiNT–mNT with the help of minor manual adjustment of binding orientations of these proteins. To obtain an optimum ternary model, an energy minimization process was performed by using the steepest descent energy minimization method. Finally, we designed a coarse-grained structural based molecular dynamics simulation experiment by taking the created unbound conformational state and the bound states obtained from the last step to show the pathway of association. The details of the construction of the ternary complex, the structure-based simulation method of the formation of the ternary complex, and the related parameters are discussed in SI Appendix and Movie S1.
Supplementary Material
Acknowledgments
O.K. acknowledges The Annual Golda Meir Fellowship and The Hebrew University Rector for the Emergency Scholarship recipients for Postdoctoral fellows, 2020–2021. H-B.M. is financed by the STIMULATE project that has received funding from the European Union’s Horizon 2020 research and innovation program under Grant Agreement 765048. This work was supported by NSF–Binational Science Foundation (BSF) Grant NSF-MCB 1613462 (to R.M.) and BSF Grant 2015831 (to R.N.), as well as by GM111364 (to R.M.). The work of J.N.O., F.B., and S.R. was supported by NSF Grants PHY-2019745 and CHE-1614101, and by Welch Foundation Grant C-1792. J.N.O. is a Cancer Prevention and Research Institute of Texas Scholar in Cancer Research. F.M. acknowledges support from the NSF Grant MCB-1943442 and the NIH Grant R35GM133631.
Footnotes
Reviewers: A.G., Los Alamos National Laboratory; M.P., University of California, Riverside; and C.W., University of Minnesota.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121491119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. Proteomics data was deposited in ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030730.
References
- 1.Braymer J. J., Lill R., Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 292, 12754–12763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boniecki M. T., Freibert S. A., Mühlenhoff U., Lill R., Cygler M., Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat. Commun. 8, 1287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lill R., Mühlenhoff U., Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77, 669–700 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Stehling O., Wilbrecht C., Lill R., Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 100, 61–77 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Lill R., Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Lill R., et al. , The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins. Eur. J. Cell Biol. 94, 280–291 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Lill R., Freibert S.-A., Mechanisms of mitochondrial iron-sulfur protein biogenesis. Annu. Rev. Biochem. 89, 471–499 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Lill R., Srinivasan V., Mühlenhoff U., The role of mitochondria in cytosolic-nuclear iron–sulfur protein biogenesis and in cellular iron regulation. Curr. Opin. Microbiol. 22, 111–119 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Karmi O., et al. , The unique fold and lability of the [2Fe-2S] clusters of NEET proteins mediate their key functions in health and disease. JBIC 23, 599–612 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamir S., et al. , Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim. Biophys. Acta 1853, 1294–1315 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Mittler R., et al. , NEET proteins: A new link between iron metabolism, reactive oxygen species, and cancer. Antioxid. Redox Signal. 30, 1083–1095 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipper C. H., et al. , Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 115, 272–277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molino D., et al. , Chemical targeting of NEET proteins reveals their function in mitochondrial morphodynamics. EMBO Rep. 21, e49019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., et al. , CISD3 inhibition drives cystine-deprivation induced ferroptosis. Cell Death Dis. 12, 839 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pila-Castellanos I., et al. , Mitochondrial morphodynamics alteration induced by influenza virus infection as a new antiviral strategy. PLoS Pathog. 17, e1009340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nechushtai R., et al. , The balancing act of NEET proteins: Iron, ROS, calcium and metabolism. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118805 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Karmi O., et al. , Interactions between mitoNEET and NAF-1 in cells. PLoS One 12, e0175796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohn Y. S., et al. , NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc. Natl. Acad. Sci. U.S.A. 110, 14676–14681 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landry A. P., Ding H., Redox control of human mitochondrial outer membrane protein MitoNEET [2Fe-2S] clusters by biological thiols and hydrogen peroxide. J. Biol. Chem. 289, 4307–4315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai F., et al. , The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 112, 3698–3703 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danielpur L., et al. , GLP-1-RA corrects mitochondrial labile iron accumulation and improves beta-cell function in type 2 Wolfram syndrome. J. Clin. Endocrinol. Metab. 101, 3592–3599 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Darash-Yahana M., et al. , Breast cancer tumorigenicity is dependent on high expression levels of NAF-1 and the lability of its Fe-S clusters. Proc. Natl. Acad. Sci. U.S.A. 113, 10890–10895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golinelli-Cohen M. P., et al. , Redox control of the human iron-sulfur repair protein MitoNEET activity via its iron-sulfur cluster. J. Biol. Chem. 291, 7583–7593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vernay A., et al. , MitoNEET-dependent formation of intermitochondrial junctions. Proc. Natl. Acad. Sci. U.S.A. 114, 8277–8282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furihata T., et al. , mitoNEET regulates mitochondrial iron homeostasis interacting with transferrin receptor. bioRxiv, [Preprint] (2018). 10.1101/330084. Accessed 20 November 2021. [DOI]
- 26.Karmi O., et al. , A combined drug treatment that reduces mitochondrial iron and reactive oxygen levels recovers insulin secretion in NAF-1-deficient pancreatic cells. Antioxidants 10, 1160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giacomello M., Pyakurel A., Glytsou C., Scorrano L., The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 21, 204–224 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Schrepfer E., Scorrano L., Mitofusins, from mitochondria to metabolism. Mol. Cell 61, 683–694 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Pernas L., Scorrano L., Mito-morphosis: Mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 78, 505–531 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Lipper C. H., et al. , Cancer-related NEET proteins transfer 2Fe-2S clusters to Anamorsin, a protein required for cytosolic iron-sulfur cluster biogenesis. PLoS One 10, e0139699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferecatu I., et al. , The diabetes drug target MitoNEET governs a novel trafficking pathway to rebuild an Fe-S cluster into cytosolic aconitase/iron regulatory protein 1. J. Biol. Chem. 289, 28070–28086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipper C. H., et al. , Redox-dependent gating of VDAC by mitoNEET. Proc. Natl. Acad. Sci. U.S.A. 116, 19924–19929 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mena N. P., Urrutia P. J., Lourido F., Carrasco C. M., Núñez M. T., Mitochondrial iron homeostasis and its dysfunctions in neurodegenerative disorders. Mitochondrion 21, 92–105 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Senyilmaz D., et al. , Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature 525, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue Y., et al. , Endoplasmic reticulum-mitochondria junction is required for iron homeostasis. J. Biol. Chem. 292, 13197–13204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey A. K., Pain J., Dancis A., Pain D., Mitochondria export iron-sulfur and sulfur intermediates to the cytoplasm for iron-sulfur cluster assembly and tRNA thiolation in yeast. J. Biol. Chem. 294, 9489–9502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maio N., Jain A., Rouault T. A., Mammalian iron-sulfur cluster biogenesis: Recent insights into the roles of frataxin, acyl carrier protein and ATPase-mediated transfer to recipient proteins. Curr. Opin. Chem. Biol. 55, 34–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao S., Hu J., Mitochondrial fusion: The machineries in and out. Trends Cell Biol. 31, 62–74 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Sabouny R., Shutt T. E., Reciprocal regulation of mitochondrial fission and fusion. Trends Biochem. Sci. 45, 564–577 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Bai F., Morcos F., Cheng R. R., Jiang H., Onuchic J. N., Elucidating the druggable interface of protein–protein interactions using fragment docking and coevolutionary analysis. Proc. Natl. Acad. Sci. U.S.A. 113, E8051–E8058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morcos F., et al. , Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 108, E1293–E1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dominguez C., Boelens R., Bonvin A. M., HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003). [DOI] [PubMed] [Google Scholar]
- 43.King S. D., et al. , The cisd gene family regulates physiological germline apoptosis through ced-13 and the canonical cell death pathway in Caenorhabditis elegans. Cell Death Differ. 26, 162–178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X.-Y., et al. , Isoliquiritigenin induces mitochondrial dysfunction and apoptosis by inhibiting mitoNEET in a reactive oxygen species-dependent manner in A375 human melanoma cells. Oxidative Med. Cellular Longevity 2019, 9817576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geldenhuys W. J., Leeper T. C., Carroll R. T., mitoNEET as a novel drug target for mitochondrial dysfunction. Drug Discov. Today 19, 1601–1606 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Holt S. H., et al. , Activation of apoptosis in NAF-1-deficient human epithelial breast cancer cells. J. Cell Sci. 129, 155–165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Divakaruni A. S., et al. , Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 547, 309–354 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Lampl T., et al. , Isolation and functional analysis of mitochondria from cultured cells and mouse tissue. J. Vis. Exp. 97, 52076 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paddock M. L., et al. , MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc. Natl. Acad. Sci. U.S.A. 104, 14342–14347 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai F., et al. , Free energy landscape for the binding process of Huperzine A to acetylcholinesterase. Proc. Natl. Acad. Sci. U.S.A. 110, 4273–4278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clementi C., Nymeyer H., Onuchic J. N., Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J. Mol. Biol. 298, 937–953 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Noel J. K., Whitford P. C., Onuchic J. N., The shadow map: A general contact definition for capturing the dynamics of biomolecular folding and function. J. Phys. Chem. B 116, 8692–8702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roy S., Jaiswar A., Sarkar R., Dynamic asymmetry exposes 2019-nCoV prefusion spike. J. Phys. Chem. Lett. 11, 7021–7027 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Honeycutt J. D., Thirumalai D., The nature of folded states of globular proteins. Biopolymers 32, 695–709 (1992). [DOI] [PubMed] [Google Scholar]
- 55.Jana B., Onuchic J. N., Strain mediated adaptation is key for myosin mechanochemistry: Discovering general rules for motor activity. PLOS Comput. Biol. 12, e1005035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noel J. K., et al. , SMOG 2: A versatile software package for generating structure-based models. PLOS Comput. Biol. 12, e1004794 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robertson M. J., Qian Y., Robinson M. C., Tirado-Rives J., Jorgensen W. L., Development and testing of the OPLS-AA/M force field for RNA. J. Chem. Theory Comput. 15, 2734–2742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darden T., York D., Pedersen L., Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Proteomics data was deposited in ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030730.