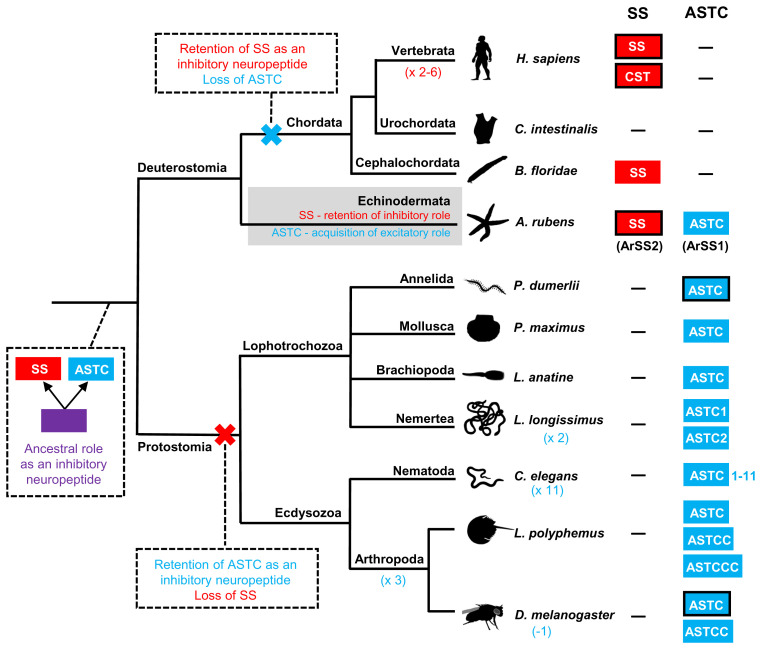
Fig. 7.
Diagram showing a model of the evolution of SS/ASTC-type neuropeptide signaling in the Bilateria, informed by the findings of this study. The occurrence of SS-type neuropeptides/precursors (red-filled boxes) and ASTC-type neuropeptide/precursors (blue-filled boxes) in different taxa is shown on the right side of the phylogenetic tree, with a dash (-) indicating absence of an SS-type and/or ASTC-type neuropeptide/precursor. Taxa in which the receptors for a SS-type or ASTC-type neuropeptide have been identified experimentally are identified by black outlining of boxes. Based on the phylogenetic distribution of SS/ASTC-type neuropeptides, it is inferred that duplication of a gene encoding an ancestral SS/ASTC-type neuropeptide (purple box) in a common ancestor of the Bilateria gave rise to genes encoding precursors of paralogous SS-type and ASTC-type neuropeptides, as shown at the root of the tree. ASTC-type and SS-type neuropeptides were lost in chordates and protostomes, respectively, because of functional redundancy as inhibitory neuropeptides. However, both ASTC-type (e.g., ArSS1) and SS-type (e.g., ArSS2) neuropeptides were retained in echinoderms because SS-type neuropeptides (e.g., ArSS2) were retained as inhibitory neuropeptides and ASTC-type neuropeptides (e.g., ArSS1) acquired a new role as excitatory neuropeptides. Lineage-specific genome/gene duplications (e.g., ×2) or gene losses (e.g., −1) that have given rise to variation in the number of SS/ASTC-type neuropeptides in different taxa are shown in parentheses. Species names are as follows: H. sapiens (Homo sapiens), C. intestinalis (Ciona intestinalis), B. floridae (Branchiostoma floridae), A. rubens (Asterias rubens), P. dumerilii (Platynereis dumerilii), P. maximus (Pecten maximus), L. anatine (Lingula anatine), L. longissimus (Lineus longissimus), C. elegans (Caenorhabditis elegans), L. polyphemus (Limulus polyphemus), and D. melanogaster (Drosophila melanogaster).