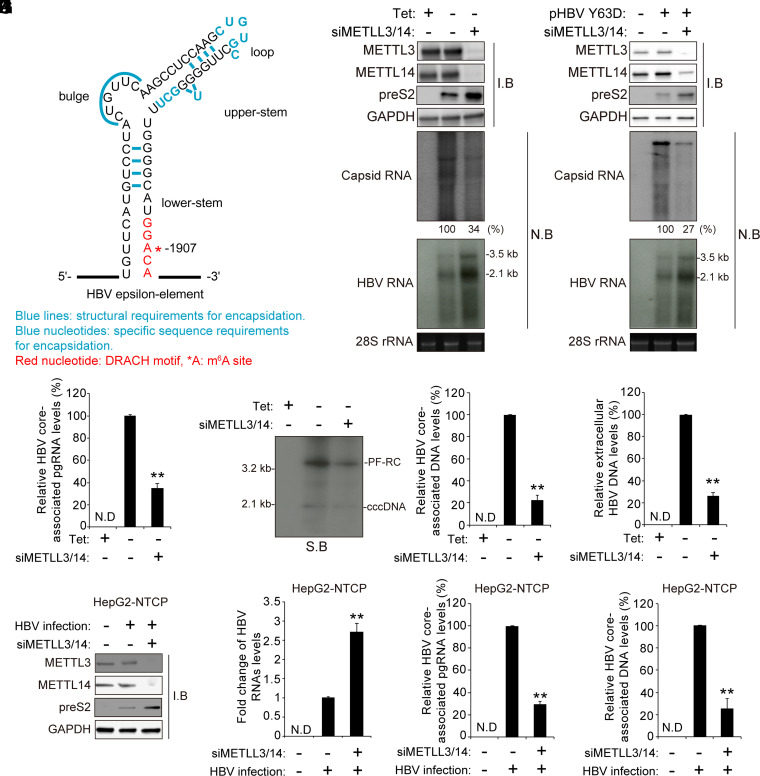
Fig. 1.
The cellular m6A methyltransferases affect the HBV pgRNA encapsidation. (A) The secondary structure of the epsilon elements is shown with the functional requirements for RNA packaging. They are represented as follows: Blue lines indicate structural requirements for encapsidation, blue nucleotides indicate specific sequence requirements for encapsidation, red nucleotides indicate DRACH motif, and *A is the m6A site. (B) HepAD38 cells stably expressing HBV were grown in the absence or presence of tetracycline for 72 h, and the cells were then transfected with the METTL3/14-specific siRNAs. After 48 h, cellular lysates, core-associated pgRNA, and total RNA were extracted from these cells for Western blotting and Northern blotting, respectively. (C) Huh7 cells were transfected with pHBV Y63D plasmid (RT-defective mutant) for 24 h, and then the siRNAs of METTL3/14 were transfected into Huh7 cells expressing pHBV Y63D plasmid. After 48 h, cellular lysates, core-associated pgRNA, and total RNA were extracted from these cells. The indicated proteins were analyzed by Western blotting. The encapsidated and cellular HBV RNA were analyzed by Northern blotting. (D–G) The siRNAs of METTL3/14 were transfected into stably expressing HBV HepAD38 cells grown in the absence or presence of tetracycline for 72 h. After 48 h, cells and supernatant were harvested. The core-associated pgRNA was analyzed by RT-qPCR (D). Hirt’s extract was prepared and subjected to Southern blot assays (E). The core-associated DNA and extracellular DNA levels were analyzed by qPCR (F and G). (H–K) HepG2-NTCP cells were infected with 2.5 × 103 genome equivalents per cell of HBV particles. After 10 d, cellular lysates, total RNA, and core-associated pgRNA and DNA were extracted from these cells. The indicated proteins were analyzed by Western blotting (H). The cellular HBV RNA and core-associated pgRNA were analyzed by RT-qPCR (I and J). The HBV core-associated DNA was assayed by qPCR (K). In E, F to G, and I to K, the error bars represent the SDs of three independent experiments. The P values are calculated via an unpaired Student’s t test. **P < 0.01. siMETTL3/14, siRNAs of METTL3/14; I.B, immunoblotting; N.B, Northern blotting; N.D, not detected.