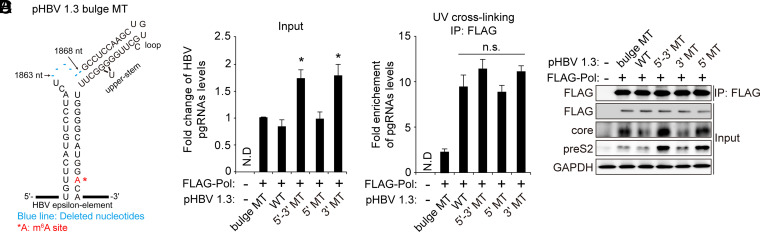
Fig. 3.
The m6A modification of the 5′ epsilon of the HBV pgRNA does not affect viral pol–5′ epsilon interaction. (A) pHBV 1.3 Bulge MT was generated by the deletion of nucleotides from 1863 to 1868 nt of pHBV 1.3-mer. They are represented as follows: Blue lines indicate the position of deleted nucleotides, and *A is the m6A site. (B–D) Huh7 cells were cotransfected with FLAG–pol and pHBV 1.3 WT or 5′-3′ MT, 5′ MT, or 3′ MT plasmids. After 48 h, cells were washed with PBS, and then cells were irradiated with UV for RNA–protein cross-linking. Total RNA and cell lysates were extracted from these cells. FLAG-tagged HBV Pol proteins were immunoprecipitated using anti-FLAG M2 magnetic beads from UV-irradiated cell lysates. Immunoprecipitated RNAs were extracted by TRIzol. The input HBV pgRNA was analyzed by RT-qPCR (B). Immunoprecipitated HBV pgRNA levels were normalized by input HBV pgRNA levels by RT-qPCR (C). Immunoprecipitated FLAG–Pol and the indicated input proteins were analyzed by Western blotting (D). In B and C, the error bars represent the SDs of three independent experiments. The P values are calculated via an unpaired Student’s t test. *P < 0.05. IP, immunoprecipitation; N.D, not detected; n.s., nonsignificant.