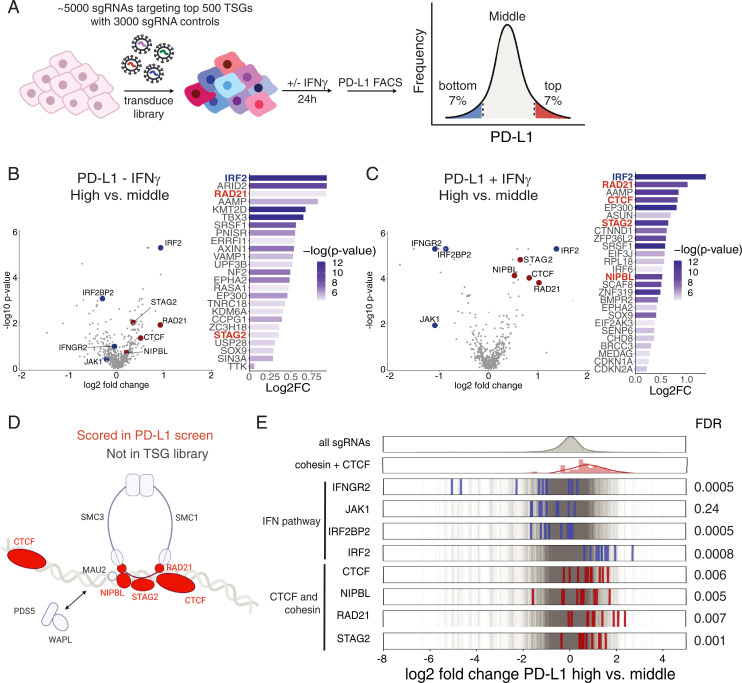
Fig. 1.
CRISPR-Cas9 screen of basal and inducible regulators of PD-L1 cell surface expression. (A) Graphical representation of the FACS-based PD-L1 screen with the TSG CRISPR-Cas9 library. (B and C) Log2 fold-changes (log2FC) and −log10(P values) for the top 25 genes with the lowest P value enriched in the high (top 7%) versus middle PD-L1 sort (middle ∼80%) in the presence and absence of IFN-γ. Volcano plots of the CRISPR-Cas9 screen of PD-L1 negative regulators in the absence and presence of IFN-γ. Log2FC and P values calculated with MAGeCK of high- versus middle-bin HMECs infected with TSG CRISPR-Cas9 library. Cohesin and CTCF genes are depicted in red, and IFN pathway genes are depicted in blue. (D) Diagram of the components of the cohesin complex and CTCF DNA-binding proteins, which were present in the TSG library (red) and scored in the FACS-based CRISPR-Cas9 screen. Created with BioRender. (E) Frequency histograms of enrichment or depletion (log2 fold-change PD-L1 sort high versus middle) of all sgRNAs in gray, IFN-γ pathway sgRNAs in blue, and cohesin and CTCF pathway sgRNAs in red.