Abstract
BACKGROUND:
Suicide is a leading cause of death worldwide, and nonfatal suicide attempts, which occur far more frequently, are a major source of disability and social and economic burden. Both have substantial genetic etiology, which is partially shared and partially distinct from that of related psychiatric disorders.
METHODS:
We conducted a genome-wide association study (GWAS) of 29,782 suicide attempt (SA) cases and 519,961 controls in the International Suicide Genetics Consortium (ISGC). The GWAS of SA was conditioned on psychiatric disorders using GWAS summary statistics via multitrait-based conditional and joint analysis, to remove genetic effects on SA mediated by psychiatric disorders. We investigated the shared and divergent genetic architectures of SA, psychiatric disorders, and other known risk factors.
RESULTS:
Two loci reached genome-wide significance for SA: the major histocompatibility complex and an intergenic locus on chromosome 7, the latter of which remained associated with SA after conditioning on psychiatric disorders and replicated in an independent cohort from the Million Veteran Program. This locus has been implicated in risk-taking behavior, smoking, and insomnia. SA showed strong genetic correlation with psychiatric disorders, particularly major depression, and also with smoking, pain, risk-taking behavior, sleep disturbances, lower educational attainment, reproductive traits, lower socioeconomic status, and poorer general health. After conditioning on psychiatric disorders, the genetic correlations between SA and psychiatric disorders decreased, whereas those with nonpsychiatric traits remained largely unchanged.
CONCLUSIONS:
Our results identify a risk locus that contributes more strongly to SA than other phenotypes and suggest a shared underlying biology between SA and known risk factors that is not mediated by psychiatric disorders.
Suicide is a worldwide public health problem, accounting for almost 800,000 deaths per year (1). Nonfatal suicide attempt (SA), defined as self-injurious behavior with the intent to die, has been estimated to occur over 20 times more frequently and is a major source of disability, reduced quality of life, and social and economic burden (1,2). The lifetime prevalence of SA in adults ranges from 0.5% to 5% worldwide (3). There are several well-established comorbidities and risk factors for SA, with psychiatric illness having the strongest effect on lifetime suicide rates (4,5). However, the vast majority of patients with psychiatric disorders never attempt suicide (6-8). Other major risk factors for SA include prior self-injurious thoughts and behaviors (9), physical illness or disability (10,11), sleep disorders (12-15), family history of psychiatric disorders (16), substance abuse (17), smoking (18-20), impulsivity (21) and social factors including childhood maltreatment (21), isolation (22), and stressful life events (23).
Both suicide and SA are heritable, with estimates from genetic epidemiology studies ranging from 17% to 55% (24-26). Several genome-wide association studies (GWASs) of SA have reported significant single nucleotide polymorphism (SNP)–heritability estimates of ~4%, indicating an underlying polygenic architecture (27-31). Using polygenic risk scoring or genetic correlation analyses, these studies have also demonstrated shared genetic etiology between SA and psychiatric disorders, with major depressive disorder (MDD) showing the largest genetic overlap (28,29,31). This genetic overlap, along with the high prevalence of MDD in the population (32), make it a particularly salient risk factor. Importantly, genetic epidemiology studies have consistently indicated a genetic component of SA that is partially distinct from that of psychiatric disorders (25). Consistent with this, one GWAS of SA that covaried for cases’ psychiatric diagnoses estimated a SNP-heritability of 1.9% (27).
With few genetic samples collected specifically for SA, studies often rely on individuals ascertained for psychiatric disorders. For example, a large GWAS of SA included over 6500 cases from clinical cohorts of MDD, bipolar disorder (BIP), and schizophrenia (SCZ) cases, within the Psychiatric Genomics Consortium (PGC) (31). In an “SA within psychiatric diagnosis” study design, SA cases were compared with cases of the same psychiatric disorder without SA, in order to disentangle the genetic etiology of SA and psychiatric disorders. While GWAS of SA have found genome-wide significant associations (27-31), thus far none of these loci have replicated, possibly owing to limited statistical power or different study designs that may probe varying components of the genetic etiology of SA. Depending on the method of ascertainment, the prevalence of psychiatric disorders may be much higher in SA cases than in controls in these studies, which may confound the genetics of SA. Well-powered and carefully designed studies are necessary to dissect the contribution of genetic variation to SA versus psychiatric disorders and advance our understanding of the genetics of SA.
Here, we present the first GWAS meta-analysis of SA from the International Suicide Genetics Consortium (ISGC), including over 29,000 SA or suicide cases from 18 cohorts worldwide. We identify novel loci implicated in SA, disentangle the genetic etiology of SA from that of MDD and psychiatric disorders, and characterize the genetic relationship among SA, psychiatric disorders, and a range of other risk factors.
METHODS AND MATERIALS
Cohorts and Case Definition
The primary SA meta-analysis comprised 18 cohorts (Table S1 in Supplement 2; Supplement 1) ascertained for psychiatric disorders, including substance use (12 cohorts), studies of suicide or SA (4 cohorts), and population-based biobanks (2 cohorts). Cases were individuals who made a nonfatal SA (16 cohorts) or died by suicide (2 cohorts). A nonfatal SA was defined as a lifetime act of deliberate self-harm with intent to die. Information on SA was ascertained using structured clinical interviews for 10 cohorts, self-report questionnaires for 4 cohorts, and hospital records or International Classification of Diseases codes for 2 cohorts. Cases of death by suicide were ascertained from the Utah State Office of the Medical Examiner or the Medical Examiner’s Office of the Hyogo Prefecture and the Division of Legal Medicine, at the Kobe University Graduate School of Medicine in Japan. A proportion of cases in the iPSYCH and Columbia University cohorts had died by suicide, determined using the Cause of Death Register in Denmark and the Columbia Classification Algorithm for Suicide Assessment, respectively (33). Individuals only endorsing suicidal ideation or nonsuicidal self-injurious behavior were not included as cases. There were 14 cohorts of European (EUR) ancestries, 2 of admixed African American (AA) ancestries, and 2 of East Asian (EAS) ancestries. All individual studies received institutional and ethical approval from their local institutional review board. Detailed cohort information is in Supplement 1 and Table S1 in Supplement 2.
Control Definition
All controls ascertained on psychiatric disorders were screened for the absence of lifetime SA. Controls from general population cohorts were screened for the absence of SA, if possible; however, because the prevalence of SA in the general population is low (3), some cohorts included unscreened controls. No controls in this study were screened for suicidal ideation or nonsuicidal self-injurious behavior. The primary SA GWAS included 29,782 cases and 519,961 controls from 18 cohorts (Table 1). Genome-wide significant associations with SA were tested in an independent replication cohort of 14,089 SA cases and 395,359 controls from Million Veteran Program (details in Supplement 1).
Table 1.
Numbers of Cases and Controls for 18 Cohorts in the International Suicide Genetics Consortium
| Cohort (Ancestry) | SA Cases | Controls |
|---|---|---|
| Psychiatric Genomics Consortium MDD (EUR) | 1528 | 16,626 |
| Psychiatric Genomics Consortium BIP (EUR) | 3214 | 17,642 |
| Psychiatric Genomics Consortium SCZ (EUR) | 1640 | 7112 |
| Psychiatric Genomics Consortium ED (EUR) | 170 | 5070 |
| Army STARRS (EUR) | 670 | 10,637 |
| German Borderline Genomics Consortium (EUR) | 481 | 1653 |
| UK Biobank (EUR) | 2433 | 334,766 |
| iPSYCH (EUR) | 7003 | 52,227 |
| Janssen (EUR) | 255 | 1684 |
| Yale-Penn (EUR) | 475 | 1817 |
| GISS Ukraine (EUR) | 660 | 660 |
| Columbia University (EUR) | 577 | 1233 |
| Australian Genetics of Depression Study and QSkin Study (EUR) | 2792 | 20,193 |
| University of Utah (EUR) | 4692 | 20,702 |
| Japan (EAS) | 746 | 14,049 |
| CONVERGE (EAS) | 1148 | 6515 |
| Grady Trauma Project (Admixed AA) | 669 | 4473 |
| Yale-Penn (Admixed AA) | 629 | 2902 |
| Total | 29,782 | 519,961 |
AA, African American; Army STARRS, Army Study to Assess Risk and Resilience in Servicemembers; BIP, bipolar disorder; EAS, East Asian; ED, eating disorder; EUR, European; GISS, Genetic Investigation of Suicide and Suicide Attempt; MDD, major depressive disorder; SA, suicide attempt; SCZ, schizophrenia.
Genotyping, Quality Control, and Imputation
Cohorts were required to have at least 200 cases prior to quality control for inclusion. Samples underwent standard genotyping, quality control, and imputation, performed by the collaborating research teams using comparable procedures (details per cohort available in Supplement 1). Briefly, samples were genotyped on microarrays, with the exception of the China, Oxford and Virginia Commonwealth University Experimental Research on Genetic Epidemiology (CONVERGE) study, which used low-coverage sequencing. Standard parameters were used to retain individuals and SNPs after quality control for missingness, relatedness, and Hardy-Weinberg equilibrium. Imputation was performed using the appropriate ancestry reference panels, resulting in >7.7 million SNPs that were well-represented across cohorts. Identical individuals between the PGC and UK Biobank cohorts were detected using genotype-based checksums (https://personal.broadinstitute.org/sripke/share_links/zpXkV8INxUg9bayDpLToG4g58TMtjN_PGC_SCZ_w3.0718d.76) and removed from PGC cohorts. There was no other known overlap of controls between any of the 18 cohorts.
GWASs and Meta-analysis
GWASs were performed in each cohort separately, and procedures are outlined in Supplement 1. GWASs were conducted within ancestry group, covarying for ancestry-informative principal components, genomic relatedness matrices, or factors capturing site of recruitment or genotyping batch, as required. The linkage disequilibrium score regression (LDSC) intercept was calculated for all GWAS results to estimate potential confounding from cryptic relatedness or population stratification (34). Studies with significant LDSC intercepts (p < .05) were corrected for confounding by multiplying the standard error per SNP by the square root of the intercept (34). A transancestry meta-analysis was conducted using an inverse variance-weighted fixed-effects model in METAL (35), implemented using the Rapid Imputation for COnsortias PIpeLIne (36). A EUR-only meta-analysis was also conducted (SA-EUR) (26,590 cases and 492,022 controls). The weighted mean allele frequency and imputation INFO score per SNP was calculated, weighted by the effective sample size per cohort. SNPs with a weighted minor allele frequency of <1%, weighted INFO score <0.6, or SNPs present in <80% of total effective sample size were removed from the meta-analysis results. A genome-wide significant locus was defined as the region around a SNP with p < 5.0 × 10−8 with linkage disequilibrium (LD) r2 > 0.1, within a 3000 kb window, based on the LD structure of the Haplotype Reference Consortium European ancestries reference panel (version 1.0) (37).
Statistical Conditioning on Psychiatric Disorders
The results of the SA-EUR meta-analysis were conditioned on the genetics of MDD using multitrait-based conditional and joint analysis using GWAS summary data (mtCOJO) (38), implemented in the GCTA software package (39). mtCOJO (38) estimates the effect size of a SNP on an outcome trait conditioned on exposure trait(s). Genome-wide significant SNPs for the exposure are used as instruments to estimate the effect of the exposure on the outcome, and this effect is used to perform genome-wide conditioning, yielding conditioned effect sizes and p values for the outcome trait. We conditioned SA (outcome) on MDD (exposure), because MDD is the most prevalent psychiatric disorder among individuals who die by suicide (40) and has the highest genetic correlation with SA (28). The SA-EUR GWAS summary statistics were used as the outcome trait, because mtCOJO requires an ancestry-matched LD reference panel and GWAS summary statistics for the exposure trait. The PGC MDD GWAS results (excluding 23andMe) (41) were used as the exposure, and the results yielded GWAS summary statistics for SA conditioned on MDD (SA-EUR∣MDD). mtCOJO is robust to sample overlap between the GWAS of the exposure and outcome. To select SNPs as instruments, independence was defined as SNPs more than 1 megabase apart or with LD r2 < 0.05 based on the 1000 Genomes Project Phase 3 EUR reference panel (42). To obtain at least 10 independent instruments for MDD, the genome-wide significance threshold was adjusted to p < 5.0 × 10−7, leading to 15 SNPs used. In a further sensitivity analysis, GWAS summary statistics for BIP (43) and SCZ (44) were additionally included as exposure traits.
LD Score Regression
LDSC (34) was used to estimate the phenotypic variance in SA explained by common SNPs (SNP-heritability, ) from GWAS summary statistics. was calculated on the liability scale assuming a lifetime prevalence of SA in the general population of 2% (middle of the range reported worldwide) (3). The bivariate genetic correlation attributable to genome-wide SNPs (rg) was estimated between the SA-EUR and SA-EUR∣ MDD GWAS and a range of psychiatric disorders, self-harm ideation, and propensity toward risk-taking behavior, using the largest available GWAS summary statistics (Bonferroni-corrected significance threshold p < .0042, adjusting for 12 traits tested). Differences in rg with SA-EUR versus SA-EUR∣ MDD were tested for deviation from 0, using the block jackknife method, implemented in LDSC software (45). The rgs of SA-EUR and SA-EUR∣MDD with 768 other nonoverlapping human diseases and traits were calculated on LD Hub (46) (Bonferroni-corrected significance threshold p < 6.51 × 10−5). Traits were precategorized manually into 15 risk factor groups previously ascribed to SA (4,5,10): autoimmune disease, neurologic disease, heart disease, hypertension, diabetes, kidney disease, cancer, alcohol use, smoking, pain, psychiatric, sleep, life stressors, socioeconomic, and education/cognition. There were 259 traits belonging to these categories, and a second reviewer validated the categories assigned to traits and their relevance to SA risk.
Polygenic Risk Scoring
Polygenic risk scores (PRSs) for SA were tested for association with SA or death by suicide versus controls in 7 target cohorts: PGC MDD, BIP and SCZ, CONVERGE (EAS ancestries), the University of Utah (suicide death cohort), Yale-Penn (AA ancestries), and Grady Trauma Project (AA ancestries). The primary SA GWAS meta-analysis was repeated excluding each cohort in turn, to create independent discovery datasets. PRSs were generated using PRS-CS (47), which uses a Bayesian regression framework to place continuous shrinkage priors on effect sizes of SNPs in the PRS, adaptive to the strength of their association signal in the discovery GWAS, and the LD structure from an external reference panel (47). The 1000 Genomes EUR, EAS, or African reference panels (42) were used to estimate LD between SNPs, as appropriate for each target cohort. PLINK 1.9 (48) was used to weight SNPs by their effect sizes calculated using PRS-CS and sum all SNPs into PRS for each individual in the target cohorts. PRSs were tested for association with case versus control status in the target cohort using a logistic regression model including covariates as per the GWAS. The amount of phenotypic variance explained by the PRS (R2) was calculated on the liability scale, assuming a lifetime prevalence of SA in the general population of 2% (3). Analyses in the PGC cohorts were repeated using PRSs generated from the SA-EUR∣MDD GWAS results, excluding each PGC cohort in turn. Analyses performed are summarized in Table S2 in Supplement 2 (Bonferroni-corrected significance threshold p < 3.12 × 10−3, adjusting for 16 tests).
RESULTS
SA Shows Significant SNP-Heritability and PRS Associations
The primary SA GWAS included 29,782 cases and 519,961 controls from 18 cohorts (Table 1). Cases were predominantly of EUR ancestries (90%), with 6% of EAS ancestries and 4% of admixed AA ancestries. Case definition was lifetime SA, with ~20% of cases having died by suicide. The SNP-heritability () SA was 6.8% (SE = 0.005, p = 2.00 × 10−42) on the liability scale. The LDSC intercept was 1.04 (SE = 0.01, p = 2.84 × 10−4), and the attenuation ratio was 0.14 (SE = 0.04), indicating that the majority of GWAS test statistic inflation was due to polygenicity (Figure S1 in Supplement 1). PRSs for SA were tested in 7 target cohorts (Table S2 in Supplement 2). SA PRSs were significantly associated with SA in the PGC MDD, BIP, and SCZ cohorts, with a phenotypic explained variance (R2) of 0.69% (p = 7.17 × 10−15), 0.68% (p = 8.11 × 10−28), and 0.88% (p = 1.24 × 10−17), respectively (liability scale). PRSs for SA were also associated with death by suicide in the University of Utah cohort, explaining slightly more phenotypic variance (R2 = 1.08%, p = 9.79 × 10−81). The rg between the University of Utah suicide death GWAS and a meta-analysis of the nonfatal SA cohorts in our study was 0.77 (SE = 0.08, p = 3.08 × 10−20). Examining the performance of SA PRSs across ancestries showed a significant association with SA in the CONVERGE EAS cohort, although with a lower explained variance (R2 = 0.25%, p = 3.06 × 10−3). Analyses in 2 admixed AA cohorts showed variable results (R2 = 0.21%, p = 5.28 × 10−1 and R2 = 0.58%, p = 3.44 × 10−3, respectively) (Table S2 in Supplement 2).
GWAS of SA Identifies Locus With Stronger Effect on SA Than Psychiatric Disorders
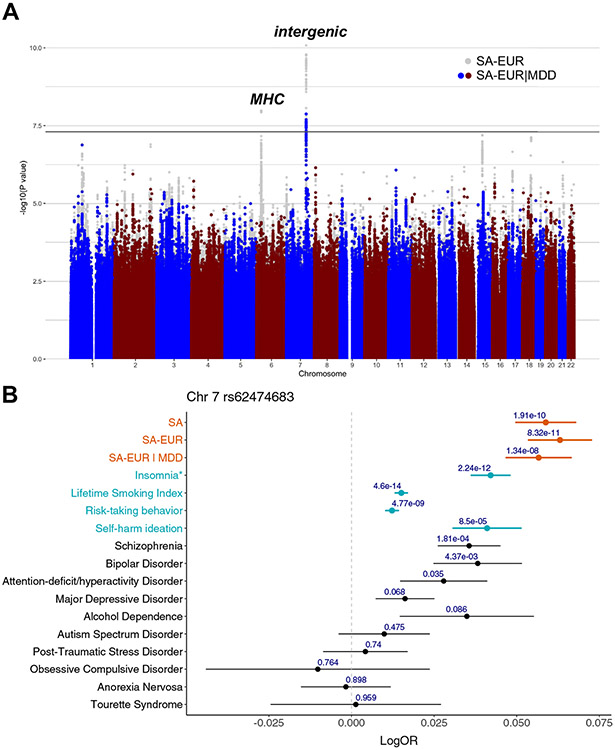
The primary SA GWAS identified 2 genome-wide significant loci (p < 5 × 10−8) (Table S3 in Supplement 2). The most strongly associated locus was in an intergenic region on chromosome 7 (index SNP rs62474683, odds ratio A allele = 1.06 [1.04–1.08], p = 1.91 × 10−10, frequency in cases = 0.52, frequency in controls = 0.50, I2 heterogeneity index = 0%) (forest plot Figure S2 in Supplement 1). The second genome-wide significant locus was in the major histocompatibility complex (MHC) (index SNP rs71557378, odds ratio T allele = 1.10 [1.06–1.13], p = 1.97 × 10−8, frequency in cases = 0.91, frequency in controls = 0.90, I2 heterogeneity index = 46%) (forest plot Figure S3 in Supplement 1). Both loci were also genome-wide significant in the SA-EUR meta-analysis, with the same effect sizes (Table S4 in Supplement 2). In order to identify SA genetic effects not mediated by MDD, we conditioned the SA-EUR GWAS on the genetic effects of MDD via mtCOJO. After conditioning, only the chromosome 7 locus remained genome-wide significant (index SNP = rs62474683, odds ratio A allele = 1.06 [1.04–1.08], p = 1.33 × 10−8) (Figure 1A). Figures S4 and S5 in Supplement 1 show regional association plots of the loci before and after conditioning. The association of the chromosome 7 index SNP with SA was further replicated in the independent Million Veteran Program cohort (rs62474683, odds ratio A allele = 1.03 [1.01–1.07], p = 3.27 × 10−3), while the index SNP in the MHC was not associated with SA in this cohort (Table S4 in Supplement 2).
Figure 1.
Genome-wide significant locus contributes to SA more strongly than psychiatric disorders and other traits. (A) Manhattan plot: the x-axis shows genomic position, and the y-axis shows statistical significance as −log10(p value). The gray points in the background depict the results of SA-EUR, and the colored points in the foreground depict the results after conditioning these results on MDD (SA-EUR∣MDD). The horizontal line shows the genome-wide significance threshold (p < 5.0 × 10−8). (B) Forest plot: the points indicate the log odds ratio of the A allele at rs62474683 (SA-index single nucleotide polymorphism on chromosome 7) on each phenotype, and the error bars show the standard error. The p value of association with each phenotype is shown above the error bars. *For insomnia, the effect size of a variant in high linkage disequilibrium with the index single nucleotide polymorphism is shown instead (rs12666306 A allele, linkage disequilibrium r2 = 0.94 with rs62474683 A allele). MDD, major depressive disorder; MHC, major histocompatibility complex; OR, odds ratio; SA, suicide attempt; SA-EUR, European-only suicide attempt meta-analysis; SA-EUR∣MDD, SA-EUR results after conditioning on MDD.
Examination of the chromosome 7 locus in published GWAS results using the Open Targets Genetics web portal (49) indicated smaller and nonsignificant effects on all psychiatric disorders (Figure 1B). Additionally, the SA-index SNP has been implicated at genome-wide significance in lifetime smoking index (50) (accounts for duration and amount of smoking) and propensity toward risk-taking behavior (51), although again with smaller effect sizes than on SA (Figure 1B; Tables S5 and S6 in Supplement 2). Pairwise GWAS analysis (see Supplement 1) of the genomic region containing the chromosome 7 locus suggested the existence of a single putative causal variant shared between SA and these phenotypes (lifetime smoking index: posterior probability = 0.99, risk-taking behavior: posterior probability = 1) (Table S7 in Supplement 2). Furthermore, a variant in high LD with the chromosome 7 index SNP (rs12666306, LD r2 = 0.94) has a positive genome-wide significant effect on insomnia (reported in GWAS catalog, full summary statistics not available) (Figure 1B; Tables S5 and S6 in Supplement 2). The SA-index SNP has also been implicated in self-harm ideation (52), although not at genome-wide significance, and with a smaller effect size than on SA (Figure 1B).
MAGMA (53) enrichment analyses performed on the primary SA GWAS (see Supplement 1) showed significant enrichment of SA associations in 7 genes (Table S8 in Supplement 2), including BTN2A1, which is a brain-expressed gene (54) located within the MHC, that encodes a plasma-membrane protein. There was no enrichment of SA association signal in any of the biological gene sets tested (Table S9 in Supplement 2) or in the set of genes expressed in any of the 54 tissues from the Genotype-Tissue Expression project (Table S10 in Supplement 2). Examining individual genes, a transcriptome-wide association study (see Supplement 1) found 5 genes for which SA risk alleles were significantly associated with brain gene expression: ERC2, RP11–266A24.1, TIAF1, BACE2, and NUFIP2 (p < 4.28 × 10−6) (Table S11 in Supplement 2). None of these genes were within genome-wide significant loci.
Evidence for Substantial Proportion of SNP-Heritability of SA Not Mediated by Psychiatric Disorders
based on the SA-EUR GWAS was 7.5% (SE = 0.006, p = 3.02 × 10−40) on the liability scale (Table S12 in Supplement 2). Conditioning SA-EUR on MDD resulted in a 45% decrease in the of SA to 4.1% (SE = 0.005, p = 1.20 × 10−16) on the liability scale (Table S12 in Supplement 2). Conditioning on BIP and SCZ in addition to MDD did not further change the estimate ( = 4.1%, SE = 0.005, p = 1.20 × 10−16). The SA-EUR∣MDD results showed comparable and complete rg with a direct GWAS of SA within psychiatric diagnosis (Supplement 1), confirming the validity of the statistical conditioning approach to control for the genetic effects of psychiatric disorders.
Significant Genetic Overlap Between SA and Psychiatric Traits or Disorders
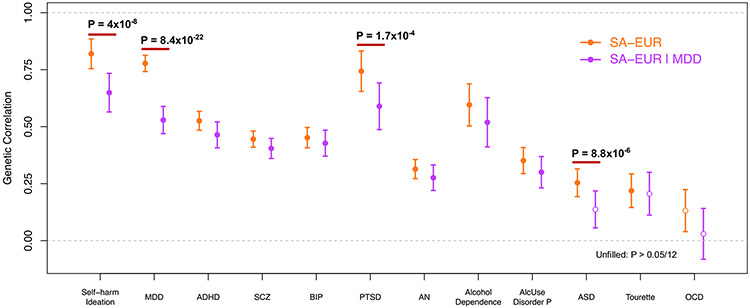
Genetic correlations were calculated to explore the genetic overlap between SA and 12 psychiatric traits or disorders before and after conditioning on MDD. The SA-EUR GWAS showed significant rg with 11 traits or disorders tested, most strongly with self-harm ideation (rg = 0.82, SE = 0.07, p = 3.57 × 10−36), MDD (rg = 0.78, SE = 0.04, p = 4.11 × 10−106), and posttraumatic stress disorder (rg = 0.74, SE = 0.09, p = 5.29 × 10−17) (Figure 2; Table S13 in Supplement 2). Moderate genetic correlations were also observed between SA and SCZ, attention-deficit/hyperactivity disorder, BIP, posttraumatic stress disorder, and alcohol dependence (rgs 0.45–0.74) (Figure 2; Table S13 in Supplement 2).
Figure 2.
Substantial genetic correlation of SA with psychiatric traits or disorders before and after conditioning SA on MDD. Unfilled points indicate genetic correlations that did not pass the Bonferroni-corrected significance threshold (p < 4.17 × 10−3). Error bars represent the standard error. p values indicate significant differences in genetic correlation after conditioning that pass Bonferroni correction. ADHD, attention-deficit/hyperactivity disorder; AlcUse Disorder P, Alcohol Use Disorders Identification Test-P (measure of problematic consequences of drinking); AN, anorexia nervosa; ASD, autism spectrum disorder; BIP, bipolar disorder; MDD, major depressive disorder; OCD, obsessive-compulsive disorder; PTSD, posttraumatic stress disorder; SA, suicide attempt; SA-EUR, European-only suicide attempt meta-analysis; SA-EUR∣MDD, SA-EUR results after conditioning on MDD; SCZ, schizophrenia.
To investigate whether these genetic correlations were mediated by MDD, we estimated rg with the same traits and disorders using the SA-EUR∣MDD results. Most genetic correlations with psychiatric disorders remained significant after conditioning, except for autism spectrum disorder and Tourette syndrome (Figure 2; Table S13 in Supplement 2). As expected, the rg with MDD significantly decreased after conditioning (p = 8.4 × 10−22 block jackknife), as did the rgs with self-harm ideation, posttraumatic stress disorder, and autism spectrum disorder (Figure 2; Table S13 in Supplement 2). The remaining psychiatric disorders did not show Bonferroni corrected significant differences in rg after conditioning on MDD. Because conditional analysis only removes SNP effects on SA mediated by MDD, the remaining rg between SA-EUR∣MDD and MDD (rg = 0.53, SE = 0.06, p = 8.9 × 10−19) indicates pleiotropic SNP effects.
Substantial Shared Genetic Architecture of SA and Nonpsychiatric Risk Factors Not Mediated by MDD
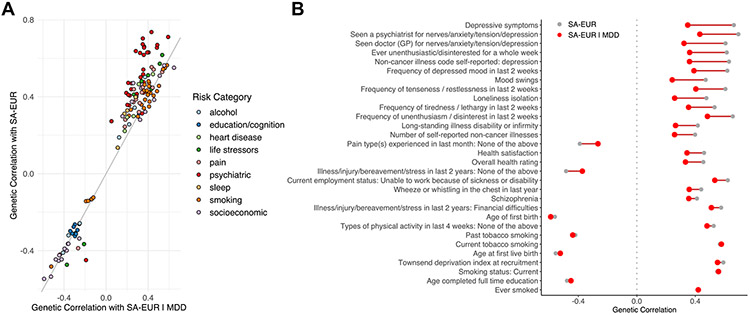
To assess the shared genetic architecture of SA, psychiatric, and nonpsychiatric phenotypes, we calculated genetic correlations of SA with 768 nonoverlapping phenotypes (46). There were 198 phenotypes that showed a significant rg with SA-EUR, 133 of which were in one of the predefined SA risk categories (Figure 3A; Table S14 in Supplement 2). The most significant genetic correlations were predominantly with traits related to depressive symptoms, smoking, and socioeconomic status. On examining phenotypes in the risk categories after conditioning on MDD, 110 phenotypes retained a significant rg with SA-EUR∣MDD (Table S14 in Supplement 2). Within the psychiatric risk category, there was a 38% average decrease in the magnitude of genetic correlations with SA-EUR after conditioning, whereas the rg values in other risk categories were much less affected by conditioning (smoking: 4.6% decrease, education/cognition: 3% decrease, alcohol: 14.5% decrease, and socioeconomic: 9.3% decrease) (Figure 3B).
Figure 3.
Conditioning SA on MDD reduces genetic correlation with psychiatric phenotypes but has limited effect on other traits. (A) Comparison of significant genetic correlations with the SA-EUR vs. genetic correlations with SA-EUR∣MDD. Data include 198 significant genetic correlations after Bonferroni correction (p < .05/768 = 6.51 × 10−5) annotated by risk category. (B) Top 30 phenotypes with the most significant genetic correlations with SA-EUR before (gray) and after (red) conditioning on MDD (SA-EUR∣MDD). Full genetic correlation results, including standard errors, are provided in Table S14 in Supplement 2. GP, general practitioner; MDD, major depressive disorder; SA, suicide attempt; SA-EUR, European-only suicide attempt meta-analysis; SA-EUR∣MDD, SA-EUR results after conditioning on major depressive disorder.
DISCUSSION
We present a GWAS of SA in over 29,000 cases, identifying 2 genome-wide significant loci, including one more strongly associated with SA than psychiatric disorders or related traits. We demonstrate that a substantial proportion of the SNP-heritability of SA is independent of psychiatric diagnosis. Finally, we show that the genetic liability to SA not mediated by psychiatric disorders is shared with the genetic architecture of nonpsychiatric risk factors.
The locus most strongly associated with SA was in an intergenic region on chromosome 7. The index SNP had a larger effect on SA than on any common psychiatric disorder, remained genome-wide significant after conditioning on MDD, and replicated in an independent cohort from the Million Veteran Program. Taken together, these results suggest that the genetic association with SA at this locus is not mediated through risk for psychiatric disorders. Functional genomic data do not clearly link this variant to any gene, with the nearest gene being a long noncoding RNA (LINC01392) 149 kb away. The index SNP (rs62474683) is a methylation quantitative trait locus, with the SA risk allele associated with decreased methylation of a nearby DNA methylation site (probe cg04544267) in blood (55). However, this methylation site has not been linked to any gene transcript. Intriguingly, SA risk alleles at this locus have been implicated at genome-wide significance in risk-taking behavior (51), smoking (50), and insomnia (56). While variants in the MHC also reached genome-wide significance for SA, this effect did not remain after conditioning on MDD, suggesting that this association may be a byproduct of psychiatric diagnosis. Indeed, variants in the MHC have previously been associated with risk for a range of psychiatric disorders, including MDD (57).
Our GWAS results provide robust evidence of the of SA, with an estimate of 6.8% on the liability scale (7.5% based on SA-EUR). Importantly, conditioning on MDD resulted in a smaller but significant estimate (4.1%), corroborating previous reports (25,27) of the independent genetic contribution to SA, and illustrating the importance of accounting for potential confounding from the genetics of psychiatric disorders. Traditionally, GWASs have sought to dissect the specific genetic component of SA by studying SA within psychiatric diagnosis or covarying for cases’ psychiatric diagnoses (27). Here, we demonstrate that statistical conditioning is an appropriate and easily applicable approach to control for the genetic effects of psychiatric disorders, producing equivalent results to a direct GWAS of SA within psychiatric diagnosis (Supplement 1).
SA showed substantial positive genetic correlation with many psychiatric disorders, the highest being with MDD (rg = 0.78, SE = 0.03), consistent with previous reports (28,29,31). Genetic overlap was also particularly strong with posttraumatic stress disorder, attention-deficit/hyperactivity disorder, SCZ, and BIP (rg = 0.44 – 0.74). After conditioning on MDD, there was a modest decrease in the genetic correlation of SA with most psychiatric disorders. Notably, SA remained strongly genetically correlated with MDD (rg = 0.53, SE = 0.06, p = 8.85 × 10−19), representing pleiotropic effects between them. This genetic correlation would only be eliminated if all SNP effects on SA were mediated by MDD. Pleiotropy between psychiatric disorders is widespread (58,59), and accordingly, genetic overlap between SA and related disorders is anticipated. Our findings suggest that many pleiotropic genetic variants increase the risk for SA directly, independent of their effects on psychiatric disorders.
Significant genetic overlap was found between SA and many nonpsychiatric traits, including smoking, lower socioeconomic status, pain, lower educational attainment, reproductive traits, risk-taking behavior, sleep disturbances, and poorer overall general health. While conditioning SA on MDD reduced genetic correlations with psychiatric disorders, the genetic correlation of SA with most nonpsychiatric traits remained unchanged. This suggests a shared genetic architecture between SA and these risk factors that is not mediated by psychiatric illness. There is substantial epidemiological literature on the relationship between sleep disorders (12-15), smoking (18-20), and socioeconomic factors (60-62) and risk for SA but less on genetic overlap between them. We have not examined potential causal relationships between these risk factors and SA, but future Mendelian randomization studies that will become possible with further increases in the power of SA GWAS may highlight modifiable risk factors.
Several limitations of our study must be noted. Cases were defined using a variety of diagnostic interviews, self-report, or hospital records, which may result in phenotypic heterogeneity. However, suicidal intent was central to all phenotype definitions, and a previous study found 98% concordance between self-report of lifetime SA and face-to-face clinician interview (63). Our GWAS included both nonfatal SA and suicide death cases, and these phenotypes were highly but imperfectly genetically correlated (rg = 0.77). Genetic correlations between SA and psychiatric disorders were examined using publicly available GWAS summary statistics; however, the prevalence of SA among the cases in these studies is unknown. Finally, population, demographic, and environmental factors are always present in genetic analyses, and while our sample is large and diverse, we did not have sufficient data to assess their possible contribution or confounding effects.
This first collaborative SA GWAS by the ISGC is almost 5-fold larger than previous studies, substantially improving statistical power. We identify a robustly associated SA risk locus and demonstrate genetic liability to SA that is not mediated through psychiatric disorders but is shared with known risk factors. We emphasize that genetic risk does not currently have meaningful predictive utility for SA, and its premature use in clinical or direct-to-consumer settings could be harmful. Future larger studies dissecting the genetic etiology of SA, psychiatric disorders, and other risk factors will provide further insights into the biological mechanisms of risk and assess potential clinical utility.
Supplementary Material
KEY RESOURCES TABLE
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | ||||
| Bacterial or Viral Strain | ||||
| Biological Sample | Human DNA samples | Many sources, all are described and cited in the Supplementary Note. Some data are available online through dbGaP or from the Haplotype Reference Consortium reference panel v1.0 (http://www.haplotype-reference-consortium.org/home) | dbGaP Psychiatric Genomics Consortium bundle phs001254 | |
| Cell Line | ||||
| Chemical Compound or Drug | ||||
| Commercial Assay Or Kit | ||||
| Deposited Data; Public Database | ||||
| Genetic Reagent | ||||
| Organism/Strain | ||||
| Peptide, Recombinant Protein | ||||
| Recombinant DNA | ||||
| Sequence-Based Reagent | ||||
| Software; Algorithm | RICOPILI software suite (version 2018_Nov_23.001) which provides wrappers for standard genetic analysis software including PLINK v1.09, and METAL (version 2011-03-25). Post-GWAS analyses were conducted using mtCOJO, MAGMA v1.08, FUSION (vOct 1, 2019), LDSC v1.0.0, PRS-CS and Pairwise GWAS. | All software is publicly available and cited in the text. | ||
| Transfected Construct | ||||
| Other | Online databases Open Targets Web Portal, LD Hub, FUMA | https://genetics.opentargets.org, http://ldsc.broadinstitute.org, https://fuma.ctglab.nl |
ACKNOWLEDGMENTS AND DISCLOSURES
Statistical analyses were carried out on the NL Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and the Mount Sinai high performance computing cluster (http://hpc.mssm.edu), which is supported by the Office of Research Infrastructure of the National Institutes of Health (Grant Nos. S10OD018522 and S10OD026880). This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. This work was funded by the National Institutes of Health (Grant Nos. R01MH116269 and R01MH121455 [to DMR]), NIGMS of the National Institutes of Health (Grant No. T32GM007347 [to JK]), and the Brain & Behavior Research Foundation (NARSAD Young Investigator Award No. 29551 [to NM]).
The content is solely the responsibility of the authors and does not necessarily represent the official views of any funding body.
We thank the participants who donated their time, life experiences, and DNA to this research, and the clinical and scientific teams that worked with them.
The International Suicide Genetics Consortium has made genome-wide summary results from this study available online (https://tinyurl.com/ISGC2021). This study included some publicly available datasets accessed through dbGaP (Psychiatric Genomics Consortium [PGC] bundle phs001254) and the Haplotype Reference Consortium reference panel v.1.0 (http://www.haplotype-reference-consortium.org/home). Databases used: Open Targets Genetics web portal (https://genetics.opentargets.org), LDHub (http://ldsc.broadinstitute.org), and FUMA (https://fuma.ctglab.nl).
In the past 3 years, RCK was a consultant for Datastat, Inc., Sage Pharmaceuticals, and Takeda. HRK and JG are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. HRK is a member of an advisory board for Dicerna Pharmaceuticals and of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last 3 years by AbbVie, Alkermes, Dicerna, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor, and Amygdala Neurosciences. DL is an employee of Janssen Research & Development, LLC, and shareholder in Johnson & Johnson, the parent company of the Janssen companies. DL declares that, except for income received from her primary employer, no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional service, and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest. MBS has in the past 3 years been a consultant for Actelion, Acadia Pharmaceuticals, Aptinyx, Bionomics, BioXcel Therapeutics, Epivario, GW Pharmaceuticals, Janssen, Jazz Pharmaceuticals, and Oxeia Biopharmaceuticals. MBS has stock options in Oxeia Biopharmaceuticals and Epivario. HJG has received travel grants and speaker honoraria from Fresenius Medical Care, Neuraxpharm, Servier, and Janssen-Cilag as well as research funding from Fresenius Medical Care. OAA is a consultant for HealthLytix and received speaker’s honorarium from Lundbeck and Sunovion. RAP is employed by and holds shares in BioMarin Pharmaceuticals. MCO and MJO are supported by a collaborative research grant from Takeda Pharmaceuticals. That support did not contribute to the work described in this manuscript. EHG has served in the speakers’ bureau and the advisory board of Takeda (former Shire do Brasil) Pharmaceutical. JAR-Q was on the speakers’ bureau and/or acted as consultant for Eli Lilly, Janssen-Cilag, Novartis, Shire, Takeda, Bial, Shionogui, Lundbeck, Almirall, Braingaze, Sincrolab, Medice, and Rubió in the last 5 years. He also received travel awards (air tickets 1 hotel) for taking part in psychiatric meetings from Janssen-Cilag, Rubió, Shire, Takeda, Shionogui, Bial, Medice, and Eli Lilly. The Department of Psychiatry chaired by him received unrestricted educational and research support from the following companies in the last 5 years: Eli Lilly, Lundbeck, Janssen-Cilag, Actelion, Shire, Ferrer, Oryzon, Roche, Psious, and Rubió. VR was on the speakers’ bureau and/or acted as consultant for Takeda and Rubió in the last 5 years. She also received travel awards (air tickets 1 hotel) for taking part in psychiatric meetings from Rubió, Shire, Takeda, and Lundbeck. MC was on the speakers’ bureau and/or acted as consultant for Janssen-Cilag in the last 5 years. He also received travel awards (air tickets 1 hotel) for taking part in psychiatric meetings from Janssen-Cilag. All other authors report no biomedical financial interests or potential conflicts of interest.
Full acknowledgments are available in Supplement 1.
Appendix
From the Department of Genetics and Genomic Sciences (NM, DPi, PS, EAS), Department of Neuroscience (PS), and Department of Psychiatry (NM, DPi, RSK, PS, EAS), Icahn School of Medicine at Mount Sinai; Department of Biostatistics (HG), Departments of Psychiatry and Radiology (JJM), and Department of Psychiatry (HG, IO), Columbia University; Department of Psychiatry (KAH), Weill Cornell Medical College; Department of Psychiatry and Behavioral Sciences (TBB, AHF), State University of New York Downstate Medical Center; Department of Psychiatry (PMr), Columbia University College of Physicians and Surgeons; Columbia University College of Physicians and Surgeons (MMW); and Division of Translational Epidemiology (MMW), New York State Psychiatric Institute, New York, New York; Division of Genetic Medicine (JK, DMR), Department of Medicine, Vanderbilt Genetics Institute, and Department of Biomedical Informatics (DMR) and Department of Psychiatry and Behavioral Sciences (DMR), Vanderbilt University Medical Center, Nashville, Tennessee; Department of Genetics and Computational Biology (AIC, MER) and Genetics and Computational Biology (SEM), QIMR Berghofer Medical Research Institute; School of Biomedical Sciences (AIC, MER), Faculty of Medicine, Institute for Molecular Bioscience (EMB, MT, NRW), Queensland Brain Institute (DM, NRW), and Centre for Children’s Health Research (EMB), The University of Queensland; School of Psychology and Counseling (DM), Queensland University of Technology, Brisbane, Queensland, Australia; National Institute for Health Research Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust (JRIC, GB); Institute of Psychiatry, Psychology and Neuroscience (JT), Department of Psychological Medicine; Institute of Psychology, Psychiatry & Neuroscience (EJSS-B); Department of Medical & Molecular Genetics (CML); and Social Genetic and Developmental Psychiatry Centre (JRIC, PMu, RAP, MRy, GB, CML), King’s College London; National Institute for Health Research Biomedical Research Centre (JT), King’s College London and South London and Maudsley National Health Service Foundation Trust; Division of Psychiatry (NB, AM, JP), University College London; and Genetics (RAP), BioMarin Pharmaceuticals; London, United Kingdom; Department of Psychiatry (ACE, ARD, TBB, AHF, KSK), Virginia Commonwealth University, Richmond, Virginia; Department of Psychiatry (DFLy, JG), Veterans Affairs Connecticut Healthcare Center, West Haven, Connecticut; Division of Human Genetics (DFLy, JG), Department of Psychiatry, Yale University School of Medicine, New Haven, Connecticut; Department of Psychiatry and Behavioral Sciences (AL, AP, EBB), Emory University School of Medicine, Atlanta, Georgia; Department of Psychiatry (ASh, EDB, ETM, HC, ARD) and Biomedical Informatics (HC), University of Utah School of Medicine, Salt Lake City, Utah; Centre for Genomics and Personalized Medicine (ASt, ADB, DD), Centre for Integrative Sequencing (ASt, PBMo, ADB, DD), Department of Biomedicine (ASt, ADB, DD), National Centre for Register-Based Research (ZY, PBMo, EA), Centre for Integrated Register-based Research (PBMo, EA), Psychosis Research Unit (OM), and Lundbeck Foundation Initiative for Integrative Psychiatric Research (ASt, ADB, AE, DD), iPSYCH, Aarhus University; and Lundbeck Foundation Initiative for Integrative Psychiatric Research (EA, DMH, OM, MN, TW), Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Aarhus, Denmark; Institute of Epidemiology and Preventive Medicine (M-HS, WJC, P-HK), College of Public Health, National Taiwan University; Department of Psychiatry (WJC, H-CC, S-CL, C-ML, P-HK), National Taiwan University Hospital; and Department of Psychiatry (H-GH), National Taiwan University Hospital and College of Medicine, Taipei, Taiwan; School of Psychology (HJW), Curtin University; and Division of Paediatrics (HJW), The University of Western Australia, Perth, Western Australia, Australia; Department of Psychiatry (HJW, LMT, ZY, CMB), Department of Genetics (ZY), and Department of Nutrition (CMB), University of North Carolina at Chapel Hill, Chapel Hill, North Carolina; Division of Psychiatry (MAd, JDH, AMM) and Institute for Genetics and Molecular Medicine (DPo), University of Edinburgh, Edinburgh, United Kingdom; Department of Psychiatry and Psychotherapy (SA, SRi), Universitätsmedizin Berlin; and Department of Psychiatry (SRo), Universitätsmedizin Berlin, Corporate Member of Freie Universität Berlin, Humboldt-Universität zu Berlin, Berlin Institute of Health, Campus Benjamin Franklin, Berlin, Germany; Department of Psychiatry and Biobehavioral Sciences (MG, MKi), Semel Institute, David Geffen School of Medicine, University of California Los Angeles; David Geffen School of Medicine (MSt), University of California Los Angeles; Department of Psychiatry and Biobehavioral Sciences (MSt), Semel Institute for Neuroscience and Human Behavior, University of California Los Angeles; and Jane and Terry Semel Institute for Neuroscience and Human Behavior (RAO), Los Angeles; Biostatistics Research Center (SJai), Herbert Wertheim School of Public Health and Human Longevity Science; Department of Psychiatry (DLB, JRK, CN), Institute for Genomic Medicine (JRK) and Department of Psychiatry and School of Public Health (MBS), University of California San Diego, La Jolla; and Department of Psychiatry (WHK) and Center for Behavioral Genomics (MTT), Department of Psychiatry, University of California San Diego, San Diego, California; Department of Psychiatry (AHi), Yokohama City University Graduate School of Medicine, Yokohama, Japan; Department of Psychiatry (SO, IO), Kobe University Graduate School of Medicine, Kobe, Japan; Stanley Center for Psychiatric Research (SRi, PHL, JWS, CCZ), Stanley Center for Psychiatric Research (KCK), and Program in Medical and Population Genetics (TE, EAS), Broad Institute, Cambridge, Massachusetts; Analytical and Translational Genetics Unit (SRi, PHL), Department of Psychiatry (KCK, JWS), Psychiatric and Neurodevelopmental Genetics Unit (PHL), and Psychiatric and Neurodevelopmental Genetics Unit (JWS), Massachusetts General Hospital; Department of Epidemiology (KCK, CCZ) and Department of Environmental Health (ARo), Harvard TH Chan School of Public Health; and Department of Health Care Policy (RCK), Harvard Medical School, Boston, Massachusetts; Population Studies Center (EBW), Center for Statistical Genetics and Department of Biostatistics (MBe, LJS), and Survery Research Center (EBW), Institute for Social Research, University of Michigan, Ann Arbor, Michigan; BioRealm, LLC (AWB), Walnut, California; Oregon Research Institute (AWB), Eugene, Oregon; Department of Psychiatry (WHB), Center for Neurobiology and Behavior, and Department of Psychiatry (DWO, HRK), Perelman School of Medicine, University of Pennsylvania; Perelman School of Medicine (HH), University of Pennsylvania; Center for Applied Genomics (XC, YG, HH, DL), Children’s Hospital of Philadelphia; and VISN 4 Mental Illness Research, Education, and Clinical Center (DWO, HRK), Corporal Michael J. Crescenz VA Medical Center, Philadelphia, Pennsylvania; Department of Psychosomatic Medicine and Psychotherapy (MBh, CS) and Department of Genetic Epidemiology in Psychiatry (SHW, LZ, FS, JCF, TGS, LS, MRe), Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Mannheim, Germany; Center for Eating Disorders at Sheppard Pratt (HB, SCa); and Department of Psychiatry and Behavioral Sciences (TGS) and Department of Mental Health (AE), Johns Hopkins University School of Medicine, Baltimore, Maryland; Center for Neuropsychiatric Research (WJC), National Health Research Institutes, Miaoli County, Taiwan; Department of Psychiatry (SCo), University of Minnesota, Minneapolis, Minnesota; Inserm U955, Institut Mondor de recherches Biomédicales, Laboratoire, Neuro-Psychiatrie Translationnelle, and Fédération Hospitalo-Universitaire de Précision Médecine en Addictologie et Psychiatrie (SJam), and Faculté de Médecine (MLe), University Paris-Est-Créteil, Créteil; Hôpital Sainte-Anne (PD, PG), GHU Paris Psychiatrie et Neurosciences; Institute of Psychiatry and Neuroscience of Paris (PD, PG, NR), INSERM U1266, Université de Paris; Department of Psychiatry and Addiction Medicine (FB, MLe), Assistance Publique Hôpitaux de Paris; Paris Bipolar and TRD Expert Centres (FB), FondaMental Foundation; UMR-S1144 Team 1: Biomarkers of relapse and therapeutic response in addiction and mood disorders (FB), INSERM; Psychiatry (FB), Université Paris Diderot; Institut de Psychiatrie (BC, M-OK), CNRS GDR 3557; Department of Evaluation, Prevention and Therapeutic innovation (BC, M-OK), GHU Paris Psychiatrie et Neurosciences; Team Pathophysiology of psychiatric diseases (BC, M-OK), Université de Paris, Institute of Psychiatry and Neuroscience of Paris, INSERM U1266; and INSERM (MLe), Paris, France; Department of Psychiatry (FF-A, SJ-M), University Hospital Bellvitge-IDIBELL and CIBEROBN; Department of Psychiatry (MSA, RB, MC, JAR-Q, MRb, VRi, CS-M, LV-R), Hospital Universitari Vall d’Hebrón; Department of Genetics, Microbiology & Statistics (MSA, MRb, CS-M), University of Barcelona; Psychiatric Genetics Unit (MSA, MC, JAR-Q, MRb, CS-M, LV-R), Group of Psychiatry, Mental Health and Addiction, Vall d’Hebron Research Institute, Universitat Autònoma de Barcelona; Department of Psychiatry and Legal Medicine (RB, MC, JAR-Q, VRi), Universitat Autònoma de Barcelona; and Center for Research in Environmental Epidemiology (MKo), Barcelona, Spain; Department of Psychiatry and Psychotherapy (MMF), Ludwig-Maximilians-University; Schön Klinik Roseneck affiliated with the Medical Faculty of the University of Munich (MMF); Department of Translational Research in Psychiatry (EBB, BM-M), Max Planck Institute of Psychiatry; Institute of Psychiatric Phenomics and Genomics (MBu, TGS), University Hospital, Ludwig-Maximilians-University; Department of Psychiatry (IG), University of Munich; Max Planck Institute of Psychiatry (MI, SK, SL); and Munich Cluster for Systems Neurology (BM-M), Munich, Germany; Department of Surgery (SG), Faculty of Medicine, Department of Psychiatry (ASK, JLK, DBW, SK, JSS, CCZ), Laboratory Medicine and Pathobiology (CCZ), and Institute of Medical Sciences (CCZ, ASK, JLK, DBW), University of Toronto; Centre for Addiction and Mental Health (ASK, JLK, SK, JSS); Department of Paediatric Laboratory Medicine (CRM) and Department of Genetics and Genomic Biology (SWS), The Hospital for Sick Children; Centre for Mental Health (DBW) and Program for Eating Disorders (DBW), University Health Network; Molecular Brain Science (JBV), Centre for Addiction and Mental Health; and Molecular Brain Science (CCZ), Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Toronto, Ontario, Canada; Department of Psychiatry and Behavioral Sciences (SJG), SUNY Upstate Medical University, Syracuse, New York; Eating Recovery Center (CJ), Denver, Colorado; Department of Psychology (PKK), Florida State University, Tallahassee, Florida; Department of Psychology (KLK), Michigan State University, Lansing, Michigan; Department of Psychiatry and Psychotherapy (KL), University Medical Center, Mainz, Germany; Department of Clinical Psychology (LL), The Chicago School of Professional Psychology, Washington, DC; Behavioral Health Services (SIS), Kaiser Permanente Washington, Seattle, Washington, DC; BESE Division (PJM), King Abdullah University of Science and Technology, Thuwal, Saudi Arabia; Department of Psychiatry (PJM, EC, GP, MP), Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland; Department of Psychiatry and Behavioral Science (JEM), University of North Dakota School of Medicine and Health Sciences, Fargo, North Dakota; HudsonAlpha Institute for Biotechnology (RMM), Huntsville, Alabama; Department of Psychology (VRo), St. Petersburg State University; and Department of Borderline Disorders and Psychotherapy (VRo), V.M. Bekhterev National Medical Research Center for Psychiatry and Neurology; Saint Petersburg, Russian Federation; National Centre for Suicide Research and Prevention of Mental Ill-Health (MSo, DW), LIME, Karolinska Institutet; Department of Clinical Neuroscience (IA), Centre for Psychiatry Research, Karolinska Institutet; Department of Clinical Neuroscience (LA), Institut of Environmental Medicine (LA), and Department of Medical Epidemiology and Biostatistics (MLa, CMB), Karolinska Institutet, Stockholm, Sweden; Department of Clinical Sciences (RA), Psychiatry, Umeå University Medical Faculty, Umeå, Sweden; Department of Psychiatric Research (IA), Diakonhjemmet Hospital; NORMENT (IA), Institute of Clinical Medicine, University of Oslo; Division of Mental Health and Addiction (OAA, IM, OBS) and Department of Medical Genetics (SD), Oslo University Hospital; NORMENT (OAA, OBS), University of Oslo; and Division of Mental Health and Addiction (IM), University of Oslo, Institute of Clinical Medicine, Oslo, Norway; Discipline of Psychiatry (TMA), University of Adelaide, Adelaide, South Australia, Australia; Department of Psychiatry (MAl), Dalhousie University, Halifax, Nova Scotia, Canada; National Institute of Mental Health (MAl), Klecany, Czech Republic; Psychiatry (AA), Berkshire Healthcare NHS Foundation Trust, Bracknell, United Kingdom; Institute of Biological Psychiatry (VA, TW), Copenhagen Mental Health Services, Copenhagen University Hospital; Lundbeck Foundation Initiative for Integrative Psychiatric Research (VA, PBMo), iPSYCH; Danish Research Institute for Suicide Prevention (AE), Mental Health Centre Copenhagen; Center for Neonatal Screening (DMH), Department for Congenital Disorders, Statens Serum Institut; Mental Health Center Copenhagen (MN), Copenhagen University Hospital; and Department of Clinical Medicine (TW), and Lundbeck Foundation GeoGenetics Centre (TW), GLOBE Institute, University of Copenhagen, Copenhagen, Denmark; Biomedical Network Research Centre on Mental Health (MSA, RB, MC, JAR-Q, MRb, VRi, CS-M), Instituto de Salud Carlos III, Madrid, Spain; Department of Psychiatry and Psychotherapy (SVA, HJG) and Institute for Community Medicine (HV), University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, Germany; Department of Psychiatry (MHA), University of Coimbra, Coimbra, Portugal; Laboratory of Developmental Psychiatry (CHDB) and ADHD Outpatient Program (EHG), Adult Division, Hospital de Clínicas de Porto Alegre; Department of Genetics (CHDB) and Department of Psychiatry (EHG), Universidade Federal do Rio Grande do Sul, Porto Alegre, Rio Grande do Sul, Brazil; Department of Psychiatry (BTB), Melbourne Medical School, University of Melbourne, Melbourne, Victoria, Australia; Department of Psychiatry (BTB) and Institute of Epidemiology and Social Medicine (KB), University of Münster, Münster, Nordrhein-Westfalen, Germany; Health Sciences Research (JMB) and Department of Psychiatry & Psychology (MF), Mayo Clinic, Rochester, Minnesota; Department of Psychiatry (MPB, WC, RSK), UMC Utrecht Hersencentrum, Utrecht, the Netherlands; School of Psychology (RB, JMG), School of Psychiatry (MJG, PBMi, GR, CSW, TWW), and School of Medical Sciences (JMF, PRS), University of New South Wales; and Neuroscience Research Australia (JMF, JMG, MJG, PRS, CSW, TWW), Sydney, New South Wales, Australia; Mental Health Unit (JAC), Department of Psychiatry, Faculty of Medicine, Granada University Hospital Complex, and Department of Psychiatry (BG), Department of Biochemistry and Molecular Biology II and Institute of Neurosciences (MRv), and Department of Nursing (EM), Faculty of Medicine and Biomedical Research Centre, University of Granada, Granada, Spain; Institute of Neuroscience and Medicine (SCi, AJF), Research Centre Jülich, Jülich, Germany; Institute of Medical Genetics and Pathology (SCi, PH) and Department of Biomedicine (SCi, PH), University Hospital Basel, Basel, Switzerland; Institute of Human Genetics (SCi, FD, AJF, SH-H, PH, MMN), University of Bonn, School of Medicine & University Hospital Bonn, Bonn, Germany; Neuropsychiatric Genetics Research Group (AC), Department of Psychiatry and Trinity Translational Medicine Institute, Trinity College Dublin, Dublin, Ireland; Medical Research Council Centre for Neuropsychiatric Genetics and Genomics (NC, MLH, IJ, MCO, MJO, JTRW), Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, United Kingdom; Department of Translational Genomics (DC), University of Southern California, Pasadena, California; NORMENT (SD), KG Jebsen Centre for Psychosis Research, Department of Clinical Science, University of Bergen, Bergen, Norway; Department of Medical & Molecular Genetics (HJE), Indiana University; and Biochemistry and Molecular Biology (HJE), Indiana University School of Medicine, Indianapolis, Indiana; Centre for Human Genetics (AJF), University of Marburg, Marburg, Germany; Department of Psychiatry and Behavioral Sciences (PVG, ARS), NorthShore University HealthSystem, Evanston, Illinois; Department of Psychiatry and Behavioral Neuroscience (PVG, ARS), University of Chicago, Chicago, Illinois; Department of Psychiatry, Psychotherapy and Psychosomatics (IG, AHa, BK, DR), Martin-Luther-University Halle-Wittenberg, Halle, Germany; Biometric Psychiatric Genetics Research Unit (MG-S), Alexandru Obregia Clinical Psychiatric Hospital, Bucharest, Romania; Mental Health Department (JG-P, FM), University Regional Hospital, Biomedicine Institute, Málaga, Spain; Psychiatry (SPH), Kaiser Permanente Northern California, San Francisco, California; Department of Psychiatry (JH), Laboratory of Psychiatric Genetics, Poznan University of Medical Sciences, Poznań, Poland; Department of Psychological Medicine (LAJ), University of Worcester, Worcester, United Kingdom; Department of Psychiatry and Neuroscience (LJ, MLa), University of Gothenburg, Gothenburg, Sweden; Psychiatry (JLa), North East London NHS Foundation Trust, Ilford, Cheshire, United Kingdom; Department of Psychiatry and Behavioral Sciences (DFLn, LMW), Stanford University, Stanford, California; Department of Human Genetics (CL), Department of Psychiatry (GT), and Department of Neurology and Neurosurgery (GAR), Faculty of Medicine, McGill University; and Montreal Neurological Institute and Hospital (CL, GAR), Montreal, Québec, Canada; Cancer Epidemiology and Prevention (JLi), Sklodowska-Curie Cancer Center and Institute of Oncology, Warsaw, Poland; Research Institute (SLM), Lindner Center of HOPE, Mason, Ohio; Department of Psychiatry (YM, BWJHP), Amsterdam UMC, Vrije Universiteit and GGZ inGeest, Amsterdam, Netherlands; Mental Health (GM), Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, NTNU; and Psychiatry (GM), St. Olavs University Hospital, Trondheim, Norway; University of Liverpool (BM-M), Liverpool, United Kingdom; Psychiatry and Human Genetics (VN), University of Pittsburgh, Pittsburgh, Pennsylvania; Psychiatry (RAO), Erasmus University Medical Center, Rotterdam, the Netherlands; College of Medicine Institute for Genomic Health (CP) and Institute for Genomic Health (CP, MTP), SUNY Downstate Medical Center College of Medicine, Brooklyn, New York; Department of Psychiatry (JBP, VW), University of Iowa, Iowa City, Iowa; Department of Psychiatry (DQ), University of Oxford; and University of Oxford (RAP), St. Edmund Hall, Oxford, United Kingdom; Department of Psychiatry (ARe), Psychosomatic Medicine and Psychotherapy, University Hospital Frankfurt, Frankfurt, Germany; Department of Physiology and Biophysics (DLR), Instituto de Ciencias Biomedicas Universidade de Sao Paulo, São Paulo, São Paulo, Brazil; Human Genetics Branch (TGS), Intramural Research Program, National Institute of Mental Health; Department of Psychiatry (RJU), Uniformed University of the Health Sciences; and Division of Cancer Epidemiology and Genetics (JS), National Cancer Institute, Bethesda, Maryland; Department of Psychiatry and Psychotherapy (TGS), University Medical Center Göttingen, Göttingen, Germany; Department of Biomedical and NeuroMotor Sciences (ASe), University of Bologna, Bologna, Italy; Menninger Department of Psychiatry and Behavioral Sciences (GS), Baylor College of Medicine, Houston, Texas; Laboratory of Neuropsychiatry (GS), IRCCS Santa Lucia Foundation, Rome, Italy; Department of Environmental Epidemiology (BŚ), Nofer Institute of Occupational Medicine, Lódz, Poland; Duke Molecular Physiology Institute (AEA-K, ERH, MAH, XQ) and Department of Psychiatry and Behavioral Sciences (JCB, NAK), Duke University Medical Center; VISN 6 Mid-Atlantic Mental Illness Research (JCB, NAK) and Cooperative Studies Program Epidemiology Center, Education, and Clinical Center (ERH), and VA Health Services Research and Development Center of Innovation to Accelerate Discovery and Practice Transformation (JHL), Durham Veterans Affairs Health Care System, Durham, North Carolina; Theoretical Division (BM), Los Alamos National Laboratory, Los Alamos National Laboratory, Los Alamos, New Mexico; Center of Mental Health Research (AE), Australian National University, Canberra, Australia; Estonian Genome Center (TE), Institute of Genomics, University of Tartu, Tartu, Estonia; Neuroscience (QSL), Janssen Research & Development, LLC, Titusville, New Jersey; Department of Genetics and Computational Biology (NGM) and Department of Population Health (CMO, DCW), QIMR Berghofer Medical Research Institute, Herston, Brisbane, Queensland, Australia; and Institute of Health and Wellbeing (DJS), University of Glasgow, Glasgow, United Kingdom.
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium
Naomi R. Wray*,1,2, Stephan Ripke*,3,4,5, Manuel Mattheisen*,6,7,8, Maciej Trzaskowski1, Enda M. Byrne1, Abdel Abdellaoui9, Mark J. Adams10, Esben Agerbo11,12,13, Tracy M. Air14, Till F.M. Andlauer15,16, Silviu-Alin Bacanu17, Marie Bækvad-Hansen13,18, Aartjan T.F. Beekman19, Tim B. Bigdeli17,20, Elisabeth B. Binder15,21, Julien Bryois22, Henriette N. Buttenschøn13,23,24, Jonas Bybjerg-Grauholm13,18, Na Cai25,26, Enrique Castelao27, Jane Hvarregaard Christensen8,13,24, Toni-Kim Clarke10, Jonathan R.I. Coleman28, Lucía Colodro-Conde29, Baptiste Couvy-Duchesne2,30, Nick Craddock31, Gregory E. Crawford32,33, Gail Davies34, Franziska Degenhardt35, Eske M. Derks29, Nese Direk36,37, Conor V. Dolan9, Erin C. Dunn38,39,40, Thalia C. Eley28, Valentina Escott-Price41, Farnush Farhadi Hassan Kiadeh42, Hilary K. Finucane43,44, Jerome C. Foo45, Andreas J. Forstner35,46,47,48, Josef Frank45, Héléna A. Gaspar28, Michael Gill49, Fernando S. Goes50, Scott D. Gordon29, Shantel Marie Weinsheimer13,54, Jürgen Wellmann101, Gonneke Willemsen9, Stephanie H. Witt45, Yang Wu1, Hualin S. Xi112, Jian Yang2,113, Futao Zhang1, Volker Arolt114, Bernhard T. Baune114,115,116, Klaus Berger101, Dorret I. Boomsma9, Sven Cichon35,47,117,118, Udo Dannlowski114, E.J.C. de Geus9,119, J. Raymond Depaulo50, Enrico Domenici120, Katharina Domschke121,122, Tõnu Esko5,78, Hans J. Grabe109, Steven P. Hamilton123, Jakob Grove8,13,24,51, Lynsey S. Hall10,52, Christine Søholm Hansen13,18, Thomas F. Hansen53,54,55, Stefan Herms35,47, Ian B. Hickie56, Per Hoffmann35,47, Georg Homuth57, Carsten Horn58, Jouke-Jan Hottenga9, David M. Hougaard13,18, David M. Howard10,28, Marcus Ising59, Rick Jansen19, Ian Jones60, Lisa A. Jones61, Eric Jorgenson62, James A. Knowles63, Isaac S. Kohane64,65,66, Julia Kraft4, Warren W. Kretzschmar67, Zoltán Kutalik68,69, Yihan Li67, Penelope A. Lind29, Donald J. MacIntyre70,71, Dean F. MacKinnon50, Robert M. Maier2, Wolfgang Maier72, Jonathan Marchini73, Hamdi Mbarek9, Patrick McGrath74, Peter McGuffin28, Sarah E. Medland29, Divya Mehta2,75, Christel M. Middeldorp9,76,77, Evelin Mihailov78, Yuri Milaneschi19, Lili Milani78, Francis M. Mondimore50, Grant W. Montgomery1, Sara Mostafavi79, 80, Niamh Mullins28, Matthias Nauck81,82, Bernard Ng80, Michel G. Nivard9, Dale R. Nyholt83, Paul F. O’Reilly28, Hogni Oskarsson84, Caroline Hayward124, Andrew C. Heath89, Kenneth S. Kendler17, Stefan Kloiber59,125,126, Glyn Lewis127, Qingqin S. Li128, Susanne Lucae59, Pamela A.F. Madden89, Patrik K. Magnusson22, Nicholas G. Martin29, Andrew M. McIntosh10,34, Andres Metspalu78,129, Ole Mors13,130, Preben Bo Mortensen11,12,13,24, Bertram Müller-Myhsok15,131,132, Merete Nordentoft13,133, Markus M. Nöthen35, Michael C. O’Donovan60, Sara A. Paciga134, Nancy L. Pedersen22, Michael J. Owen60, Jodie N. Painter29, Carsten Bøcker Pedersen11,12,13, Marianne Giørtz Pedersen11,12,13, Roseann E. Peterson17,85, Wouter J. Peyrot19, Giorgio Pistis27, Danielle Posthuma86,87, Jorge A. Quiroz88, Per Qvist8,13,24, John P. Rice89, Brien P. Riley17, Margarita Rivera28,90, Saira Saeed Mirza36, Robert Schoevers91, Eva C. Schulte92,93, Ling Shen62, Jianxin Shi94, Stanley I. Shyn95, Engilbert Sigurdsson96, Grant C.B. Sinnamon97, Johannes H. Smit19, Daniel J. Smith98, Hreinn Stefansson99, Stacy Steinberg99, Fabian Streit45, Jana Strohmaier45, Katherine E. Tansey100, Henning Teismann101, Alexander Teumer102, Wesley Thompson13,54,103,104, Pippa A. Thomson105, Thorgeir E. Thorgeirsson99, Matthew Traylor106, Jens Treutlein45, Vassily Trubetskoy4, André G. Uitterlinden107, Daniel Umbricht108, Sandra Van der Auwera109, Albert M. van Hemert110, Alexander Viktorin22, Peter M. Visscher1,2, Yunpeng Wang13,54,104, Bradley T. Webb111, Brenda W.J.H. Penninx19, Roy H. Perlis38,135, David J. Porteous105, James B. Potash136, Martin Preisig27, Marcella Rietschel45, Catherine Schaefer62, Thomas G. Schulze45,93,137,138,139, Jordan W. Smoller38,39,40, Kari Stefansson99,140, Henning Tiemeier36,141,142, Rudolf Uher143, Henry Völzke102, Myrna M. Weissman74,144, Thomas Werge13,54,145, Cathryn M. Lewis*,28,146, Douglas F. Levinson*,147, Gerome Breen*,28,148, Anders D. Børglum*,8,13,24, and Patrick F. Sullivan*,22,149,150 See Supplement 1 for all affiliations of consortium members.
Bipolar Disorder Working Group of the Psychiatric Genomics Consortium
Niamh Mullins1,2, Andreas J. Forstner3,4,5, Kevin S. O’Connell6,7, Brandon Coombes8, Jonathan R.I. Coleman9,10, Zhen Qiao11, Thomas D. Als12,13,14, Tim B. Bigdeli15,16, Sigrid Børte17,18,19, Julien Bryois20, Alexander W. Charney2, Ole Kristian Drange21,22, Michael J. Gandal23, Saskia P. Hagenaars9,10, Masashi Ikeda24, Nolan Kamitaki25,26, Minsoo Kim23, Kristi Krebs27, Georgia Panagiotaropoulou28, Brian M. Schilder1,29,30,31, Laura G. Sloofman1, Stacy Steinberg32, Vassily Trubetskoy28, Bendik S. Winsvold19,33, Hong-Hee Won34, Liliya Abramova35, Kristina Adorjan36,37, Esben Agerbo14,38,39, Mariam Al Eissa40, Diego Albani41, Ney Alliey-Rodriguez42,43, Adebayo Anjorin44, Verneri Antilla45, Anastasia Antoniou46, Swapnil Awasthi28, Ji Hyun Baek47, Marie Bækvad-Hansen14,48, Nicholas Bass40, Michael Bauer49, Eva C. Beins3, Sarah E. Bergen20, Armin Birner50, Carsten Bøcker Pedersen14,38,39, Erlend Bøen51, Marco P. Boks52, Rosa Bosch53,54,55,56, Murielle Brum57, Ben M. Brumpton19, Nathalie Brunkhorst-Kanaan57, Monika Budde36, Jonas Bybjerg-Grauholm14,48, William Byerley58, Murray Cairns59, Miquel Casas53,54,55,56, Pablo Cervantes60, Toni-Kim Clarke61, Cristiana Cruceanu60,62, Alfredo Cuellar-Barboza63,64, Julie Cunningham65, David Curtis66,67, Piotr M. Czerski68, Anders M. Dale69, Nina Dalkner50, Friederike S. David3, Franziska Degenhardt3,70, Srdjan Djurovic71,72, Amanda L. Dobbyn1,2, Athanassios Douzenis46, Torbjørn Elvsåshagen18,73,74, Valentina Escott-Price75, I. Nicol Ferrier76, Alessia Fiorentino40, Tatiana M. Foroud77, Liz Forty75, Josef Frank78, Oleksandr Frei6,18, Nelson B. Freimer23,79, Louise Frisén80, Katrin Gade36,81, Julie Garnham82, Joel Gelernter83,84,85, Marianne Giørtz Pedersen14,38,39, Ian R. Gizer86, Scott D. Gordon87, Katherine Gordon-Smith88, Tiffany A. Greenwood89, Jakob Grove12,13,14,90, José Guzman-Parra91, Kyooseob Ha92, Magnus Haraldsson93, Martin Hautzinger94, Urs Heilbronner36, Dennis Hellgren20, Stefan Herms3,95,96, Per Hoffmann3,95,96, Peter A. Holmans75, Laura Huckins1,2, Stéphane Jamain97,98, Jessica S. Johnson1,2, Janos L. Kalman36,37,99, Yoichiro Kamatani100,101, James L. Kennedy102,103,104,105, Sarah Kittel-Schneider57,106, James A. Knowles107,108, Manolis Kogevinas109, Maria Koromina110, Thorsten M. Kranz57, Henry R. Kranzler111,112, Michiaki Kubo113, Ralph Kupka114,115,116, Steven A. Kushner117, Catharina Lavebratt118,119, Jacob Lawrence120, Markus Leber121, Heon-Jeong Lee122, Phil H. Lee123, Shawn E. Levy124, Catrin Lewis75, Calwing Liao125,126, Susanne Lucae62, Martin Lundberg118,119, Donald J. MacIntyre127, Sigurdur H. Magnusson32, Wolfgang Maier128, Adam Maihofer89, Dolores Malaspina1,2, Eirini Maratou129, Lina Martinsson80, Manuel Mattheisen12,13,14,106,130, Nathaniel W. McGregor131, Peter McGuffin9, James D. McKay132, Helena Medeiros108, Sarah E Medland87, Vincent Millischer118,119, Grant W. Montgomery11, Jennifer L. Moran25,133, Derek W. Morris134, Thomas W. Mühleisen4,95, Niamh O’Brien40, Claire O’Donovan82, Loes M. Olde Loohuis23,79, Lilijana Oruc135, Sergi Papiol36,37, Antonio F. Pardiñas75, Amy Perry88, Andrea Pfennig49, Evgenia Porichi46, James B. Potash136, Digby Quested137,138, Towfique Raj1,29,30,31, Mark H. Rapaport139, J. Raymond DePaulo136, Eline J. Regeer140, John P. Rice141, Fabio Rivas91, Margarita Rivera142,143, Julian Roth106, Panos Roussos1,2,29, Douglas M. Ruderfer144, Cristina Sánchez-Mora53,54,56,145, Eva C. Schulte36,37, Fanny Senner36,37, Sally Sharp40, Paul D. Shilling89, Engilbert Sigurdsson93,146, Lea Sirignano78, Claire Slaney82, Olav B. Smeland6,7, Daniel J. Smith147, Janet L. Sobell148, Christine Søholm Hansen14,48, Maria Soler Artigas53,54,56,145, Anne T. Spijker149, Dan J. Stein150, John S. Strauss102, Beata Świątkowska151, Chikashi Terao101, Thorgeir E. Thorgeirsson32, Claudio Toma152,153,154, Paul Tooney59, Evangelia-Eirini Tsermpini110, Marquis P. Vawter155, Helmut Vedder156, James T.R. Walters75, Stephanie H. Witt78, Simon Xi157, Wei Xu158, Jessica Mei Kay Yang75, Allan H. Young159,160, Hannah Young1, Peter P. Zandi136, Hang Zhou83,84, Lea Zillich78, HUNT All-In Psychiatry161, Rolf Adolfsson162, Ingrid Agartz51,130,163, Martin Alda82,164, Lars Alfredsson165, Gulja Babadjanova166, Lena Backlund118,119, Bernhard T. Baune167,168,169, Frank Bellivier170,171, Susanne Bengesser50, Wade H. Berrettini172, Douglas H.R. Blackwood61, Michael Boehnke173, Anders D. Børglum14,174,175, Gerome Breen9,10, Vaughan J. Carr176, Stanley Catts177, Aiden Corvin178, Nicholas Craddock75, Udo Dannlowski167, Dimitris Dikeos179, Tõnu Esko26,27,180,181, Bruno Etain170,171, Panagiotis Ferentinos9,46, Mark Frye64, Janice M. Fullerton152,153, Micha Gawlik106, Elliot S. Gershon42,182, Fernando S. Goes136, Melissa J. Green152,176, Maria Grigoroiu-Serbanescu183, Joanna Hauser68, Frans Henskens59, Jan Hillert80, Kyung Sue Hong47, David M. Hougaard14,48, Christina M. Hultman20, Kristian Hveem19,184, Nakao Iwata24, Assen V. Jablensky185, Ian Jones75, Lisa A Jones88, René S. Kahn2,52, John R. Kelsoe89, George Kirov75, Mikael Landén20,186, Marion Leboyer97,98,187, Cathryn M. Lewis9,10,188, Qingqin S. Li189, Jolanta Lissowska190, Christine Lochner191, Carmel Loughland59, Nicholas G. Martin87,192, Carol A. Mathews193, Fermin Mayoral91, Susan L. McElroy194, Andrew M. McIntosh127,195, Francis J. McMahon196, Ingrid Melle6,197, Patricia Michie59, Lili Milani27, Philip B. Mitchell176, Gunnar Morken21,198, Ole Mors14,199, Preben Bo Mortensen12,14,38,39, Bryan Mowry177, Bertram Müller-Myhsok62,200,201, Richard M. Myers124, Benjamin M. Neale25,45,180, Caroline M. Nievergelt89,202, Merete Nordentoft14,203, Markus M. Nöthen3, Michael C. O’Donovan75, Ketil J. Oedegaard204,205, Tomas Olsson206, Michael J. Owen75, Sara A. Paciga207, Chris Pantelis208, Carlos Pato108, Michele T. Pato108, George P. Patrinos110,209,210, Roy H. Perlis211,212, Danielle Posthuma213,214, Josep Antoni Ramos-Quiroga53,54,55,56, Andreas Reif57, Eva Z. Reininghaus50, Marta Ribasés53,54,56,145, Marcella Rietschel78, Stephan Ripke25,28,45, Guy A. Rouleau126,215, Takeo Saito24, Ulrich Schall59, Martin Schalling118,119, Peter R. Schofield152,153, Thomas G. Schulze36,78,81,136,216, Laura J. Scott173, Rodney J. Scott59, Alessandro Serretti217, Cynthia Shannon Weickert152,176,218, Jordan W. Smoller25,133,219, Hreinn Stefansson32, Kari Stefansson32,220, Eystein Stordal221,222, Fabian Streit78, Patrick F. Sullivan20,223,224, Gustavo Turecki225, Arne E. Vaaler226, Eduard Vieta227, John B. Vincent102, Irwin D. Waldman228, Thomas W. Weickert152,176,218, Thomas Werge14,229,230,231, Naomi R. Wray11,232, John-Anker Zwart18,19,33, Joanna M. Biernacka8,64, John I. Nurnberger233, Sven Cichon3,4,95,96, Howard J. Edenberg77,234, Eli A. Stahl1,2,180, Andrew McQuillin40, Arianna Di Florio75,224, Roel A. Ophoff23,79,117,235, and Ole A. Andreassen6,7
See Supplement 1 for all affiliations of consortium members.
Eating Disorders Working Group of the Psychiatric Genomics Consortium
Roger A.H. Adan1,2,3, Lars Alfredsson4, Tetsuya Ando5, Ole A. Andreassen6, Harald Aschauer7, Jessica H. Baker8, Vladimir Bencko9, Andrew W. Bergen10,11, Wade H. Berrettini12, Andreas Birgegård13,14,15, Joseph M. Boden16, Ilka Boehm17, Claudette Boni18, Vesna Boraska Perica19,20, Harry Brandt21, Gerome Breen22,23, Julien Bryois15, Katharina Buehren24, Cynthia M. Bulik8,15,25, Roland Burghardt26, Laura Carlberg27, Matteo Cassina28, Sven Cichon29,30,31, Maurizio Clementi28, Jonathan R.I. Coleman22,23, Roger D. Cone32, Philippe Courtet33, Steven Crawford21, Scott Crow34, James J. Crowley13,35, Unna N. Danner2, Oliver S.P. Davis36,37, Martina de Zwaan38, George Dedoussis39, Daniela Degortes40, Janiece E. DeSocio41, Danielle M. Dick42,43,44, Dimitris Dikeos45, Christian Dina46, Monika Dmitrzak-Weglarz47, Elisa Docampo Martinez48,49,50, Laramie E. Duncan51, Karin Egberts52, Christian R. Marshall126, Nicholas G. Martin72, Manuel Mattheisen13,14,75,127, Morten Mattingsdal6, Sara McDevitt128,129, Peter McGuffin22, Sarah E. Medland72, Andres Metspalu53,130, Ingrid Meulenbelt131, Nadia Micali132,133, James Mitchell134, Karen Mitchell135,136, Palmiero Monteleone137, Alessio Maria Monteleone124, Grant W. Montgomery72,86,138, Preben Bo Mortensen76,114,115, Melissa A. Munn-Chernoff8, Benedetta Nacmias139, Marie Navratilova63, Ioanna Ntalla39, Catherine M. Olsen140, Roel A. Ophoff141,142, Julie K. O’Toole143, Leonid Padyukov110, Aarno Palotie54,102,144, Jacques Pantel18, Hana Papezova97, Richard Parker72, John F. Pearson145, Nancy L. Pedersen15, Stefan Ehrlich17, Geòrgia Escaramís48,49,50, Tõnu Esko53,54, Thomas Espeseth55, Xavier Estivill48,49,50,56, Anne Farmer22, Angela Favaro40, Fernando Fernández-Aranda57,58, Manfred M. Fichter59,60, Krista Fischer53, James A.B. Floyd61, Manuel Föcker62, Lenka Foretova63, Andreas J. Forstner30,64,65,66, Monica Forzan28, Christopher S. Franklin19, Steven Gallinger67, Giovanni Gambaro68, Héléna A. Gaspar22,23, Ina Giegling69, Johanna Giuranna70, Paola Giusti-Rodríquez35, Fragiskos Gonidakis71, Scott Gordon72, Philip Gorwood73,74, Monica Gratacos Mayora48,49,50, Jakob Grove75,76,77,78, Sébastien Guillaume33, Yiran Guo79, Hakon Hakonarson79,80, Katherine A. Halmi81, Ken B. Hanscombe82, Konstantinos Hatzikotoulas19,83, Joanna Hauser84, Johannes Hebebrand70, Sietske G. Helder22,85, Anjali K. Henders86, Stefan Herms29,30, Beate Herpertz-Dahlmann24, Wolfgang Herzog87, Anke Hinney70, L. John Horwood16, Christopher Hübel15,22, Liselotte V. Petersen76,114,115, Dalila Pinto88, Kirstin L. Purves22, Anu Raevuori101, Nicolas Ramoz18, Ted Reichborn-Kjennerud112,146, Valdo Ricca147, Samuli Ripatti148, Stephan Ripke149,150,151, Franziska Ritschel17,152, Marion Roberts22, Dan Rujescu69, Filip Rybakowski154, Paolo Santonastaso155, André Scherag156, Stephen W. Scherer157,158, Ulrike Schmidt22, Nicholas J. Schork159, Alexandra Schosser160, Jochen Seitz24, Lenka Slachtova161, P. Eline Slagboom131, Margarita C.T. Slof-Op ‘t Landt162,163, Agnieszka Slopien164, Nicole Soranzo19,165,166,167, Sandro Sorbi139,168, Lorraine Southam19, Vidar W. Steen169,170, Michael Strober171,172, Laura M. Huckins88, James I. Hudson89, Hartmut Imgart90, Hidetoshi Inoko91, Vladimir Janout92, Susana Jiménez-Murcia57,58, Craig Johnson93, Jennifer Jordan94,95, Antonio Julià96, Gursharan Kalsi22, Deborah Kaminská97, Allan S. Kaplan98,99,100, Jaakko Kaprio101,102, Leila Karhunen103, Andreas Karwautz104, Martien J.H. Kas1,105, Walter H. Kaye106, James L. Kennedy98,99,100, Martin A. Kennedy107, Anna Keski-Rahkonen101, Kirsty Kiezebrink108, Youl-Ri Kim109, Katherine M. Kirk72, Lars Klareskog110, Kelly L. Klump111, Gun Peggy S. Knudsen112, Mikael Landén15,113, Janne T. Larsen76,114,115, Stephanie Le Hellard116,117,118, Virpi M. Leppä15, Dong Li79, Paul Lichtenstein15, Lisa Lilenfeld119, Bochao Danae Lin1, Jolanta Lissowska120, Astri Lundervold121, Jurjen Luykx1, Pierre J. Magistretti122,123, Mario Maj124, Katrin Mannik53,125, Sara Marsal96, Garret D. Stuber8,173, Patrick F. Sullivan8,15,35, Beata Świątkowska174, Jin P. Szatkiewicz35, Ioanna Tachmazidou19, Elena Tenconi40, Laura M. Thornton8, Alfonso Tortorella175,176, Federica Tozzi177, Janet Treasure22, Artemis Tsitsika178, Marta Tyszkiewicz-Nwafor164, Konstantinos Tziouvas179, Annemarie A. van Elburg2,180, Eric F. van Furth162,163, Tracey D. Wade181, Gudrun Wagner104, Esther Walton17, Hunna J. Watson8,182,183, Thomas Werge184, David C. Whiteman140, H. Erich Wichmann185, Elisabeth Widen102, D. Blake Woodside99,100,186,187, Shuyang Yao15, Zeynep Yilmaz8,35, Eleftheria Zeggini19,83, Stephanie Zerwas8, and Stephan Zipfel188
See Supplement 1 for all affiliations of consortium members.
German Borderline Genomics Consortium
Stephanie H. Witt1, Fabian Streit1, Martin Jungkunz2,3, Josef Frank1, Swapnil Awasthi4, Jens Treutlein1, Lydie Dietl5, Cornelia E Schwarze6, Norbert Dahmen8, Björn H. Schott4,9, Markus M. Nöthen10,11, Stephan Ripke4,12,13, Arian Mobascher8, Dan Rujescu7, Klaus Lieb8, Stefan Roepke5, Christian Schmahl2, Martin Bohus3, and Marcella Rietschel1
See Supplement 1 for all affiliations of consortium members.
MVP Suicide Exemplar Workgroup
Silvia Crivelli, Ph.D. (Lawrence Berkeley National Laboratory), Michelle F. Dennis, B.A. (Durham Veterans Affairs Health Care System & Duke University School of Medicine), Phillip D. Harvey, Ph.D. (University of Miami Miller School of Medicine, Miami, FL), Bruce W. Carter (VA Medical Center), Jennifer E. Huffman, Ph.D. (Massachusetts Veterans Epidemiology Research and Information Center, VA Boston Healthcare System), Daniel Jacobson, Ph.D. (Oak Ridge National Laboratory), Ravi Madduri, Ph.D. (Argonne National Laboratory), Maren K. Olsen, Ph.D. (Duke University School of Medicine), and John Pestian, Ph.D. (Oak Ridge National Laboratory).
VA Million Veteran Program (MVP)
J. Michael Gaziano, M.D., M.P.H. (co-chair, VA Boston Healthcare System), Sumitra Muralidhar, Ph.D. (co-chair, U.S. Department of Veterans Affairs), Rachel Ramoni, D.M.D., Sc.D. (U.S. Department of Veterans Affairs), Jean Beckham, Ph.D. (Durham VA Medical Center), Kyong-Mi Chang, M.D. (Philadelphia VA Medical Center), Christopher J. O’Donnell, M.D., M.P.H. (VA Boston Healthcare System), Philip S. Tsao, Ph.D. (VA Palo Alto Health Care System), James Breeling, M.D. (Ex-Officio, U.S. Department of Veterans Affairs), Grant Huang, Ph.D. (Ex-Officio, U.S. Department of Veterans Affairs), and J.P. Casas Romero, M.D., Ph.D. (Ex-Officio, VA Boston Healthcare System). MVP Program Office: Sumitra Muralidhar, Ph.D., and Jennifer Moser, Ph.D., both of U.S. Department of Veterans Affairs. MVP Recruitment/Enrollment: Recruitment/Enrollment Director/Deputy Director, Boston—Stacey B. Whitbourne, Ph.D., Jessica V. Brewer, M.P.H. (VA Boston Healthcare System). MVP Coordinating Centers: Clinical Epidemiology Research Center (CERC), West Haven—Mihaela Aslan, Ph.D. (West Haven VA Medical Center). Cooperative Studies Program Clinical Research Pharmacy Coordinating Center, Albuquerque—Todd Connor, Pharm.D., Dean P. Argyres, B.S., M.S. (New Mexico VA Health Care System). Genomics Coordinating Center, Palo Alto—Philip S. Tsao, Ph.D. (VA Palo Alto Health Care System). MVP Boston Coordinating Center, Boston—J. Michael Gaziano, M.D., M.P.H. (VA Boston Healthcare System). MVP Information Center, Canandaigua—Brady Stephens, M.S. (Canandaigua VA Medical Center). VA Central Biorepository, Boston—Mary T. Brophy, M.D., M.P.H., Donald E. Humphries, Ph.D., Luis E. Selva, Ph.D. (VA Boston Healthcare System). MVP Informatics, Boston—Nhan Do, M.D., Shahpoor Shayan (VA Boston Healthcare System). MVP Data Operations/Analytics, Boston—Kelly Cho, Ph.D. (VA Boston Healthcare System). MVP Science: Science Operations—Christopher J. O’Donnell, M.D., M.P.H. (VA Boston Healthcare System). Genomics Core— Christopher J. O’Donnell, M.D., M.P.H., Saiju Pyarajan, Ph.D. (VA Boston Healthcare System), Philip S. Tsao, Ph.D. (VA Palo Alto Health Care System). Phenomics Core—Kelly Cho, M.P.H., Ph.D. (VA Boston Healthcare System). Data and Computational Sciences—Saiju Pyarajan, Ph.D. (VA Boston Healthcare System). Statistical Genetics—Elizabeth Hauser, Ph.D. (Durham VA Medical Center). Yan Sun, Ph.D. (Atlanta VA Medical Center). Hongyu Zhao, Ph.D. (West Haven VA Medical Center. Current MVP Local Site Investigators: Peter Wilson, M.D. (Atlanta VA Medical Center); Rachel McArdle, Ph.D. (Bay Pines VA Healthcare System); Louis Dellitalia, M.D. (Birmingham VA Medical Center); Kristin Mattocks, Ph.D., M.P.H. (Central Western Massachusetts Healthcare System); John Harley, M.D., Ph.D. (Cincinnati VA Medical Center); Clement J. Zablocki (VA Medical Center); Jeffrey Whittle, M.D., M.P.H.; Frank Jacono, M.D. (VA Northeast Ohio Healthcare System); Jean Beckham, Ph.D. (Durham VA Medical Center); Edith Nourse Rogers Memorial Veterans Hospital; Salvador Gutierrez, M.D. (Edward Hines, Jr. VA Medical Center); Gretchen Gibson, D.D.S., M.P.H. (Veterans Health Care System of the Ozarks); Kimberly Hammer, Ph.D. (Fargo VA Health Care System); Laurence Kaminsky, Ph.D. (VA Health Care Upstate New York); Gerardo Villareal, M.D. (New Mexico VA Health Care System); Scott Kinlay, M.B.B.S., Ph.D. (VA Boston Healthcare System); Junzhe Xu, M.D. (VA Western New York Healthcare System); Mark Hamner, M.D. (Ralph H. Johnson VA Medical Center); Roy Mathew, M.D. (Columbia VA Health Care System); Sujata Bhushan, M.D. (VA North Texas Health Care System); Pran Iruvanti, DO, Ph.D. (Hampton VA Medical Center); Michael Godschalk, M.D. (Richmond VA Medical Center); Zuhair Ballas, M.D. (Iowa City VA Health Care System); Douglas Ivins, M.D. (Eastern Oklahoma VA Health Care System); Stephen Mastorides, M.D. (James A. Haley Veterans’ Hospital); Jonathan Moorman, M.D., Ph.D. (James H. Quillen VA Medical Center); Saib Gappy, M.D. (John D. Dingell VA Medical Center); Jon Klein, M.D., Ph.D. (Louisville VA Medical Center); Nora Ratcliffe, M.D. (Manchester VA Medical Center); Hermes Florez, M.D., Ph.D. (Miami VA Health Care System); Olaoluwa Okusaga, M.D. (Michael E. DeBakey VA Medical Center); Maureen Murdoch, M.D., M.P.H. (Minneapolis VA Health Care System); Peruvemba Sriram, M.D. (N FL/S GA Veterans Health System); Shing Shing Yeh, Ph.D., M.D. (Northport VA Medical Center); Neeraj Tandon, M.D. (Overton Brooks VA Medical Center); Darshana Jhala, M.D. (Philadelphia VA Medical Center); Samuel Aguayo, M.D. (Phoenix VA Health Care System); David Cohen, M.D. (Portland VA Medical Center); Satish Sharma, M.D. (Providence VA Medical Center); Suthat Liangpunsakul, M.D., M.P.H. (Richard Roudebush VA Medical Center); Kris Ann Oursler, M.D. (Salem VA Medical Center); Mary Whooley, M.D. (San Francisco VA Health Care System); Sunil Ahuja, M.D. (South Texas Veterans Health Care System); Joseph Constans, Ph.D. (Southeast Louisiana Veterans Health Care System); Paul Meyer, M.D., Ph.D. (Southern Arizona VA Health Care System); Jennifer Greco, M.D. (Sioux Falls VA Health Care System); Michael Rauchman, M.D. (St. Louis VA Health Care System); Richard Servatius, Ph.D. (Syracuse VA Medical Center); Melinda Gaddy, Ph.D. (VA Eastern Kansas Health Care System); Agnes Wallbom, M.D., M.S. (VA Greater Los Angeles Health Care System); Timothy Morgan, M.D. (VA Long Beach Healthcare System); Todd Stapley, D.O. (VA Maine Healthcare System); Scott Sherman, M.D., M.P.H. (VA New York Harbor Healthcare System); George Ross, M.D. (VA Pacific Islands Health Care System); Philip Tsao, Ph.D. (VA Palo Alto Health Care System); Patrick Strollo Jr., M.D. (VA Pittsburgh Health Care System); Edward Boyko, M.D. (VA Puget Sound Health Care System); Laurence Meyer, M.D., Ph.D. (VA Salt Lake City Health Care System); Samir Gupta, M.D., M.S.C.S. (VA San Diego Healthcare System); Mostaqul Huq, Pharm.D., Ph.D. (VA Sierra Nevada Health Care System); Joseph Fayad, M.D. (VA Southern Nevada Healthcare System); Adriana Hung, M.D., M.P.H. (VA Tennessee Valley Healthcare System); Jack Lichy, M.D., Ph.D. (Washington, DC VA Medical Center); Robin Hurley, M.D. (W.G., Bill Hefner VA Medical Center); Brooks Robey, M.D. (White River Junction VA Medical Center); and Robert Striker, M.D., Ph.D. (William S. Middleton Memorial Veterans Hospital).
Footnotes
Supplementary material cited in this article is available online at http://doi.org/10.1016/j.biopsych.2021.05.029.
REFERENCES
- 1.World Health Organization: Suicide data Available at: http://www.who.int/mental_health/prevention/suicide/suicideprevent/en/. Accessed May 9, 2020. [Google Scholar]
- 2.Centers for Disease Control and Prevention: WISQARS™ — Webbased injury statistics query and reporting system Available at: https://www.cdc.gov/injury/wisqars/index.html. Accessed May 9, 2020. [Google Scholar]
- 3.Nock MK, Borges G, Bromet EJ, Alonso J, Angermeyer M, Beautrais A, et al. (2008): Cross-national prevalence and risk factors for suicidal ideation, plans and attempts. Br J Psychiatry 192:98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franklin JC, Ribeiro JD, Fox KR, Bentley KH, Kleiman EM, Huang X, et al. (2017): Risk factors for suicidal thoughts and behaviors: A meta-analysis of 50 years of research. Psychol Bull 143:187–232. [DOI] [PubMed] [Google Scholar]
- 5.Fazel S, Runeson B (2020): Suicide. N Engl J Med 382:266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arsenault-Lapierre G, Kim C, Turecki G (2004): Psychiatric diagnoses in 3275 suicides: A meta-analysis. BMC Psychiatry 4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanagh JTO, Carson AJ, Sharpe M, Lawrie SM (2003): Psychological autopsy studies of suicide: A systematic review. Psychol Med 33:395–405. [DOI] [PubMed] [Google Scholar]
- 8.Qin P (2011): The impact of psychiatric illness on suicide: Differences by diagnosis of disorders and by sex and age of subjects. J Psychiatr Res 45:1445–1452. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro JD, Franklin JC, Fox KR, Bentley KH, Kleiman EM, Chang BP, Nock MK (2016): Self-injurious thoughts and behaviors as risk factors for future suicide ideation, attempts, and death: A meta-analysis of longitudinal studies. Psychol Med 46:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmedani BK, Peterson EL, Hu Y, Rossom RC, Lynch F, Lu CY, et al. (2017): Major physical health conditions and risk of suicide. Am J Prev Med 53:308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khazem LR (2018): Physical disability and suicide: Recent advancements in understanding and future directions for consideration. Curr Opin Psychol 22:18–22. [DOI] [PubMed] [Google Scholar]
- 12.Bernert RA, Nadorff MR (2015): Sleep disturbances and suicide risk. Sleep Med Clin 10:35–39. [DOI] [PubMed] [Google Scholar]
- 13.Woznica AA, Carney CE, Kuo JR, Moss TG (2015): The insomnia and suicide link: Toward an enhanced understanding of this relationship. Sleep Med Rev 22:37–46. [DOI] [PubMed] [Google Scholar]
- 14.Pigeon WR, Pinquart M, Conner K (2012): Meta-analysis of sleep disturbance and suicidal thoughts and behaviors. J Clin Psychiatry 73:e1160–e1167. [DOI] [PubMed] [Google Scholar]
- 15.Bishop TM, Walsh PG, Ashrafioun L, Lavigne JE, Pigeon WR (2020): Sleep, suicide behaviors, and the protective role of sleep medicine. Sleep Med 66:264–270. [DOI] [PubMed] [Google Scholar]
- 16.Qin P, Agerbo E, Mortensen PB (2002): Suicide risk in relation to family history of completed suicide and psychiatric disorders: A nested case-control study based on longitudinal registers. Lancet 360:1126–1130. [DOI] [PubMed] [Google Scholar]
- 17.Chesney E, Goodwin GM, Fazel S (2014): Risks of all-cause and suicide mortality in mental disorders: A meta-review. World Psychiatry 13:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poorolajal J, Darvishi N (2016): Smoking and suicide: A meta-analysis. PLoS One 11:e0156348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orri M, Séguin JR, Castellanos-Ryan N, Tremblay RE, Côté SM, Turecki G, Geoffroy MC (2021): A genetically informed study on the association of cannabis, alcohol, and tobacco smoking with suicide attempt. Mol Psychiatry 26:5061–5070. [DOI] [PubMed] [Google Scholar]
- 20.Harrison R, Munafò MR, Davey Smith G, Wootton RE (2020): Examining the effect of smoking on suicidal ideation and attempts: Triangulation of epidemiological approaches. Br J Psychiatry 217:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon K, Hoertel N, Olfson M, Wall M, Wang S, Blanco C (2018): Childhood maltreatment and impulsivity as predictors of interpersonal violence, self-injury and suicide attempts: A national study. Psychiatry Res 269:386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calati R, Ferrari C, Brittner M, Oasi O, Olié E, Carvalho AF, Courtet P (2019): Suicidal thoughts and behaviors and social isolation: A narrative review of the literature. J Affect Disord 245:653–667. [DOI] [PubMed] [Google Scholar]
- 23.Liu RT, Miller I (2014): Life events and suicidal ideation and behavior: A systematic review. Clin Psychol Rev 34:181–192. [DOI] [PubMed] [Google Scholar]
- 24.Voracek M, Loibl LM (2007): Genetics of suicide: A systematic review of twin studies. Wien Klin Wochenschr 119:463–475. [DOI] [PubMed] [Google Scholar]
- 25.Brent DA, Melhem N (2008): Familial transmission of suicidal behavior. Psychiatr Clin North Am 31:157–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Q, Heath AC, Bucholz KK, Nelson EC, Glowinski AL, Goldberg J, et al. (2002): A twin study of genetic and environmental influences on suicidality in men. Psychol Med 32:11–24. [DOI] [PubMed] [Google Scholar]
- 27.Erlangsen A, Appadurai V, Wang Y, Turecki G, Mors O, Werge T, et al. (2020): Genetics of suicide attempts in individuals with and without mental disorders: A population-based genome-wide association study. Mol Psychiatry 25:2410–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruderfer DM, Walsh CG, Aguirre MW, Tanigawa Y, Ribeiro JD, Franklin JC, Rivas MA (2020): Significant shared heritability underlies suicide attempt and clinically predicted probability of attempting suicide. Mol Psychiatry 25:2422–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strawbridge RJ, Ward J, Ferguson A, Graham N, Shaw RJ, Cullen B, et al. (2019): Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. EBioMedicine 41:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Docherty AR, Shabalin AA, DiBlasi E, Monson E, Mullins N, Adkins DE, et al. (2020): Genome-wide association study of suicide death and polygenic prediction of clinical antecedents. Am J Psychiatry 177:917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullins N, Bigdeli TB, Børglum AD, Coleman JRI, Demontis D, Mehta D, et al. (2019): GWAS of suicide attempt in psychiatric disorders and association with major depression polygenic risk scores. Am J Psychiatry 176:651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessler RC, Bromet EJ (2013): The epidemiology of depression across cultures. Annu Rev Public Health 34:119–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posner K, Oquendo MA, Gould M, Stanley B, Davies M (2007): Columbia Classification Algorithm of Suicide Assessment (C-CASA): Classification of suicidal events in the FDA’s pediatric suicidal risk analysis of antidepressants. Am J Psychiatry 164:1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. (2015): LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer CJ, Li Y, Abecasis GR (2010): METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam M, Awasthi S, Watson HJ, Goldstein J, Panagiotaropoulou G, Trubetskoy V, et al. (2020): RICOPILI: Rapid imputation for COnsortias PIpeLIne. Bioinformatics 36:930–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. (2016): A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, et al. (2018): Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Lee SH, Goddard ME, Visscher PM (2011): GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachmann S (2018): Epidemiology of suicide and the psychiatric perspective. Int J Environ Res Public Health 15:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. (2018): Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. (2015): A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. (2019): Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 51:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. (2018): Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50:381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hübel C, Gaspar HA, Coleman JRI, Finucane H, Purves KL, Hanscombe KB, et al. (2019): Genomics of body fat percentage may contribute to sex bias in anorexia nervosa. Am J Med Genet B Neuropsychiatr Genet 180:428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng J, Mesut Erzurumluoglu A, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. (2017): LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33:272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW (2019): Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun 10:1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015): Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mountjoy E, Schmidt EM, Carmona M, Schwartzentruber J, Peat G, Miranda A, et al. (2021): Open Targets Genetics: An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nature Genetics 53:1527–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wootton RE, Richmond RC, Stuijfzand BG, Lawn RB, Sallis HM, Taylor GMJ, et al. (2020): Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: A Mendelian randomisation study. Psychol Med 50:2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. (2019): Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet 51:245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campos AI, Verweij KJH, Statham DJ, Madden PAF, Maciejewski DF, Davis KAS, et al. (2020): Genetic aetiology of self-harm ideation and behaviour. Sci Rep 10:9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015): MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput Biol 11: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, et al. (2017): Genetic effects on gene expression across human tissues [published correction appears in Nature 2018; 553:530]. Nature 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Min JL, Hemani G, Hannon E, Dekkers KF, Castillo-Fernandez J, Luijk R, et al. (2021): Genomic and phenomic insights from an atlas of genetic effects on DNA methylation. Nature Genetics 53:1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jansen PR, Watanabe K, Stringer S, Skene N, Bryois J, Hammerschlag AR, et al. (2019): Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet 51:394–403. [DOI] [PubMed] [Google Scholar]
- 57.Bergen SE, O’Dushlaine CT, Ripke S, Lee PH, Ruderfer DM, Akterin S, et al. (2012): Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry 17:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brainstorm Consortium, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. (2018): Analysis of shared heritability in common disorders of the brain. Science 360:eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cross-Disorder Group of the Psychiatric Genomics Consortium (2019): Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179:1469–1482.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agerbo E, Nordentoft M, Mortensen PB (2002): Familial, psychiatric, and socioeconomic risk factors for suicide in young people: Nested case-control study. BMJ 325:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qin P, Agerbo E, Mortensen PB (2003): Suicide risk in relation to socioeconomic, demographic, psychiatric, and familial factors: A national register–based study of all suicides in Denmark, 1981–1997. Am J Psychiatry 160:765–772. [DOI] [PubMed] [Google Scholar]
- 62.Lorant V, de Gelder R, Kapadia D, Borrell C, Kalediene R, Kovács K, et al. (2018): Socioeconomic inequalities in suicide in Europe: The widening gap. Br J Psychiatry 212:356–361. [DOI] [PubMed] [Google Scholar]
- 63.Kaplan ML, Asnis GM, Sanderson WC, Keswani L, De Lecuona JM, Joseph S (1994): Suicide assessment: Clinical interview vs. self-report. J Clin Psychol 50:294–298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.