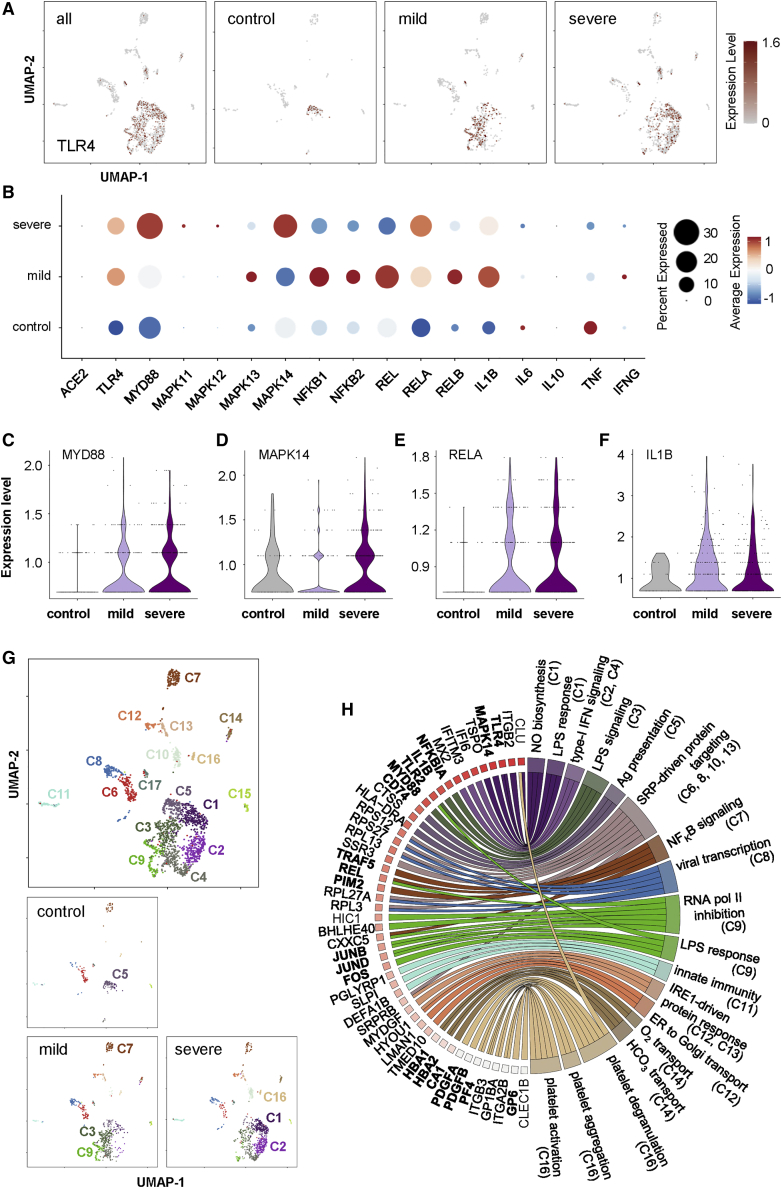
Figure 6.
Single-cell transcriptomic profiling reveals inflammatory activation of the TLR4/p38 MAPK/NF-κB/IL1B axis in human circulating ECs of COVID-19 patients
(A) Visualization of scaled expression of TLR4 in CD31+CD45− circulating ECs (2,236 out of 139,848 PBMCs) using UMAP; 50 samples from 8 mild, 9 severe COVID-19 patients, and 13 controls recruited in the same cohort of a previous study (Schulte-Schrepping et al., 2020). (B) Dot plot representation of scaled expression of ACE2, marker genes of TLR4/p38 MAPK/NF-κB and pro-inflammatory cytokines expressed in circulating ECs of all samples. (C–F) Violin plots showing expression of (C) MYD88, (D) MAPK14, (E) RELA, and (F) IL1B in circulating ECs of all samples. (G) UMAP visualization of 17 clusters of circulating ECs identified in all samples, cells are colored according to the identified clusters. (H) Circular gene ontology (GO) plot representing a selection of one to three pathways of each cluster determined by GO associated to genes detected as significantly upregulated (p < 0.05) comparing expression levels of that gene with the rest of clusters, differences were determined by Seurat’s implementation of the Wilcoxon rank-sum test.