Figure 2: Single Molecule Localization Microscopy and analysis.
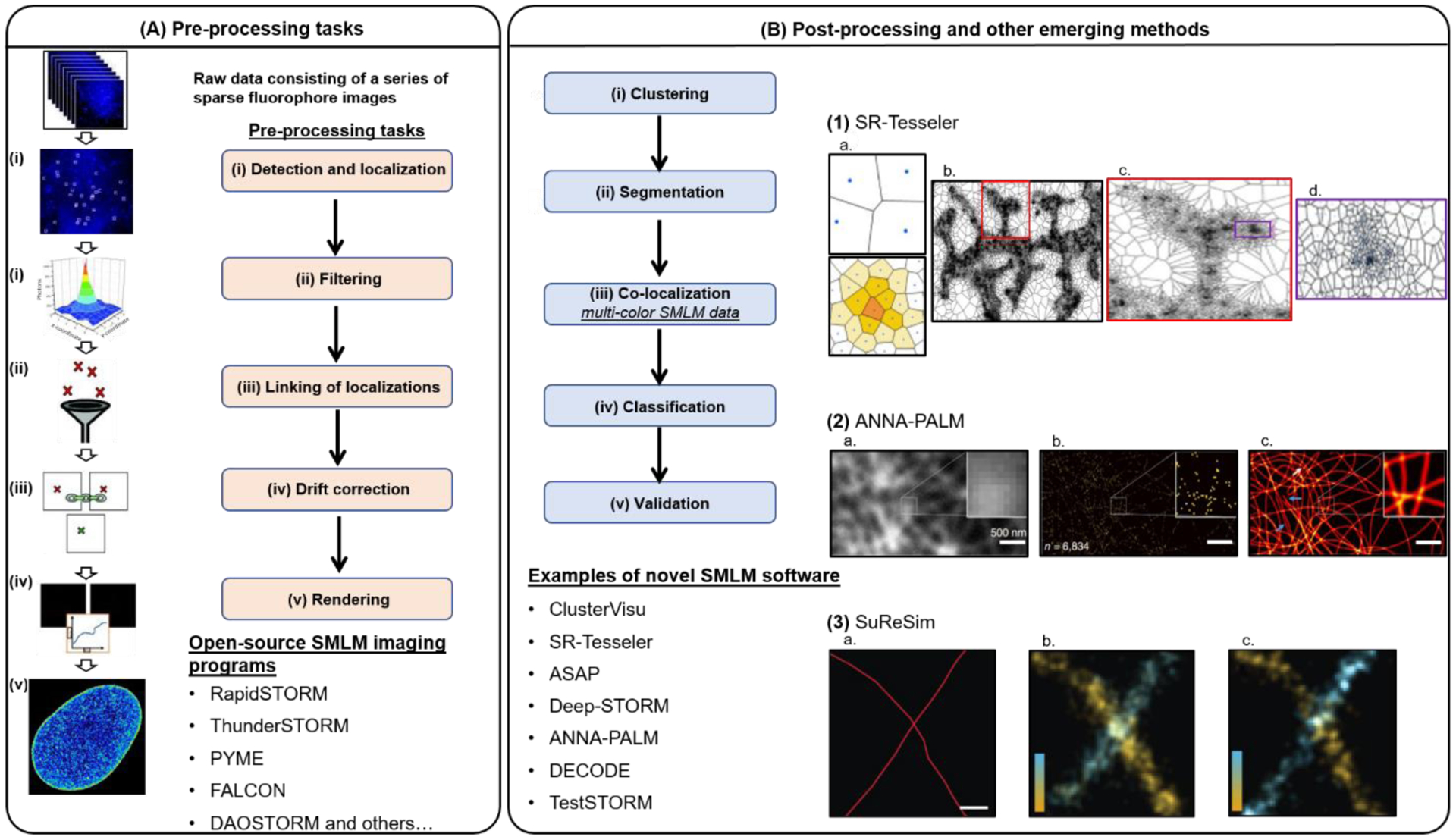
(A) The pre-processing tasks of SMLM data consist of: (i) peak detection and localization, (ii) linking of localizations, (iii) filtering, (iv) drift correction, and (v) rendering. Currently, there are more than 25 open-source software packages that can perform all or some steps of these tasks. Figure was adapted from (Brede and Lakadamyali, 2012) (B) Post-processing of SMLM images include: (i) clustering, (ii) segmentation, (iii) co-localization, (iv) classification, and (v) validation. (1) SR-Tesseler, developed by (Levet et al., 2015), segments SMLM images consisting of a wide-range of shapes and sizes using Voronoi Tessellation (1a), such as entire neuronal processes (1b, c) or nano-clusters (1d). (2) An up-and-coming category of SMLM analysis platforms are those that apply machine- and deep-learning algorithms. For example: ANNA-PALM, developed by (Ouyang et al., 2018), reconstructs super-resolution images (2c) from sparse, rapidly acquired SMLM data (2a,b). (3) Simulation of realistic SMLM data can be used to validate and compare the performance of available SMLM analysis platforms. For example, SuReSim, developed by (Venkataramani et al., 2016), is a software that can simulate biological structures under different experimental conditions; 3a shows the ground truth model, 3b shows the simulated image, and 3c shows the real-experimental data for a pair of crossed-microtubules labeled with antibodies.
