Extended Data Fig. 1. Study Design and Database Generation.
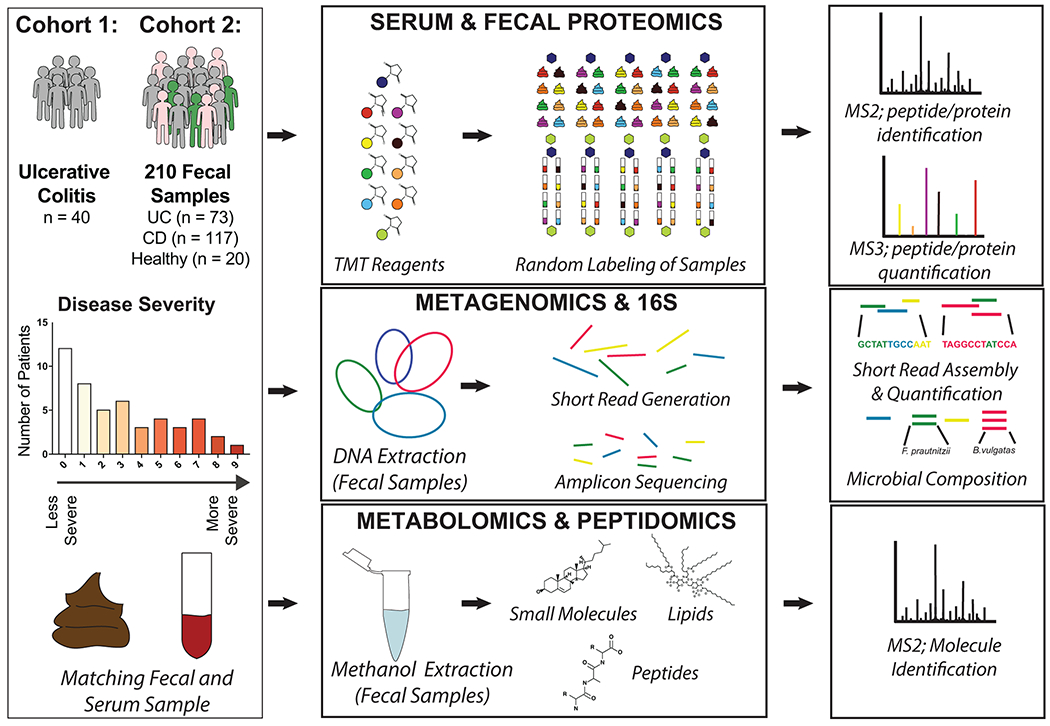
Paired fecal and serum samples were collected from 40 patients with varying severity of Ulcerative Colitis. A separately analyzed cohort of fecal samples was also collected on 210 samples with 73 UC, 117 CD and 20 healthy controls. Samples were processed for proteomics using a Tandem Mass Tag multiplexing workflow. Fecal samples were also subjected to both 16S and shotgun metagenomic analyses for microbial composition and gene quantification respectively. In parallel, a metabolomics workflow was performed on fecal samples where collected MS2 spectra were analyzed for both metabolites and peptides in two separate computational pipelines. A custom database was compiled from the metagenome of fecal samples to mediate a comparative analysis between shotgun metagenomic and metaproteomic data sets. This eliminated database dependent bias and the shared reference was used for estimating copy number.
