Fig. 5 |. DNA star design.
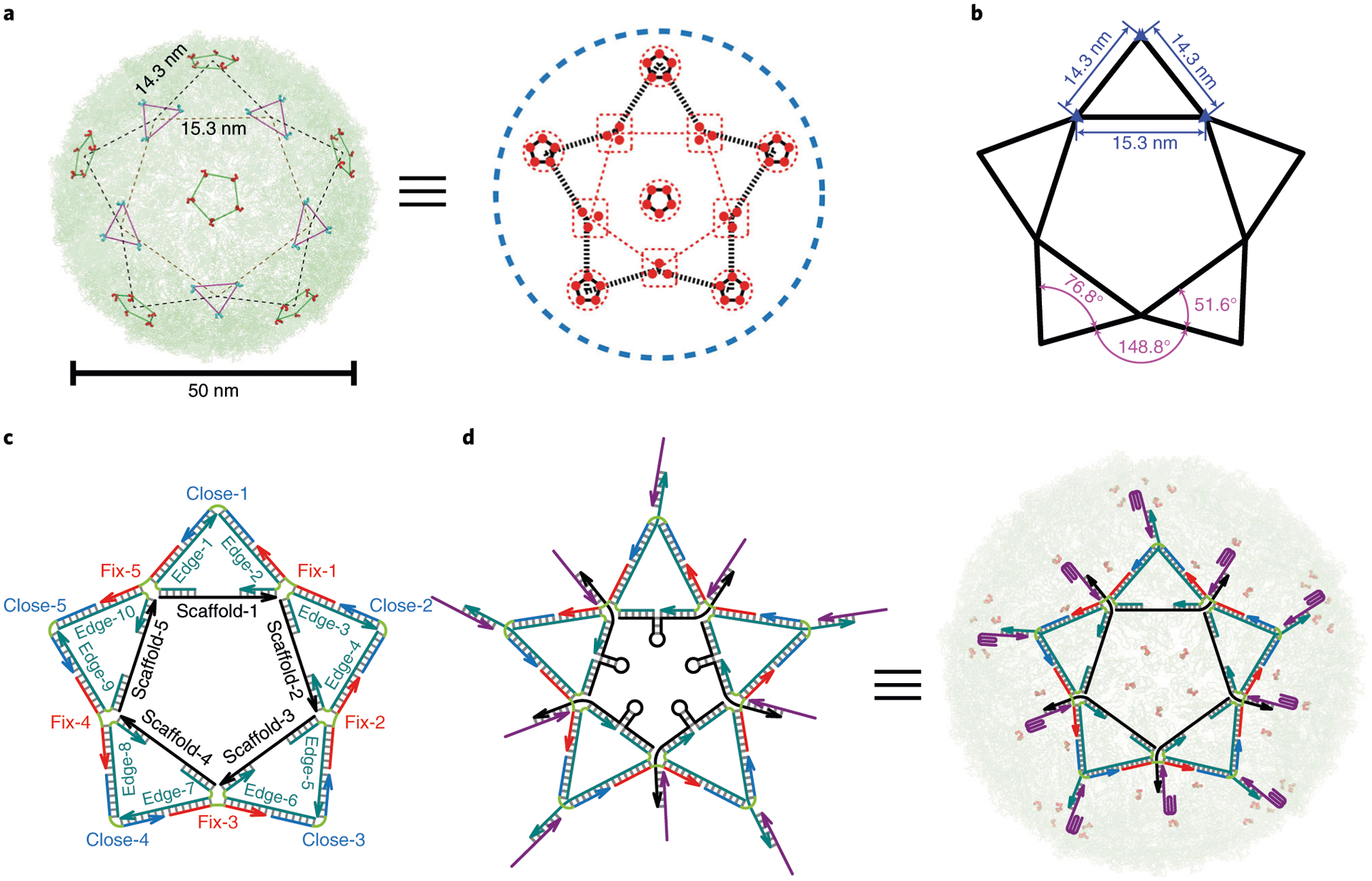
a, Distribution of DENV ED3 trivalent and pentavalent clusters presents a shape of five-pointed star7. b, Orthodromic distance between trivalent–trivalent or trivalent–pentavalent clusters is 15.3 nm or 14.3 nm. The angle of external edge–external edge (inside of an isosceles triangle), external edge–external edge (between two adjacent isosceles triangles) or external edge–internal edge is 76.8°, 148.8° or 51.6°. c, The DNA star structure consists of five ‘scaffold’ strands (S-1 to S-5) that form the pentagon inner edges, ten ‘edge’ strands that connect internal and external edges, five ‘fix’ strands that connect the external edges, and ‘close’ strands that cap the external edges of the triangle. A single-stranded region of each scaffold strand forms a hairpin loop. The sequence for each DNA strand was programmed in SEQUIN. d, 3′ overhangs on the DNA star allow aptamer incorporation7. The ten incorporated aptamers match the pattern and spacing of ED3 clusters. When bound to a virion, each of the five hairpins is stretched to a distance that fits the spacing between adjacent trivalent ED3 clusters. The switchable DNA star can be turned into a virus inhibitor or sensor (when fluorescence components are added to report virus binding). a and d adapted with permission from ref. 7, Springer Nature Ltd.
