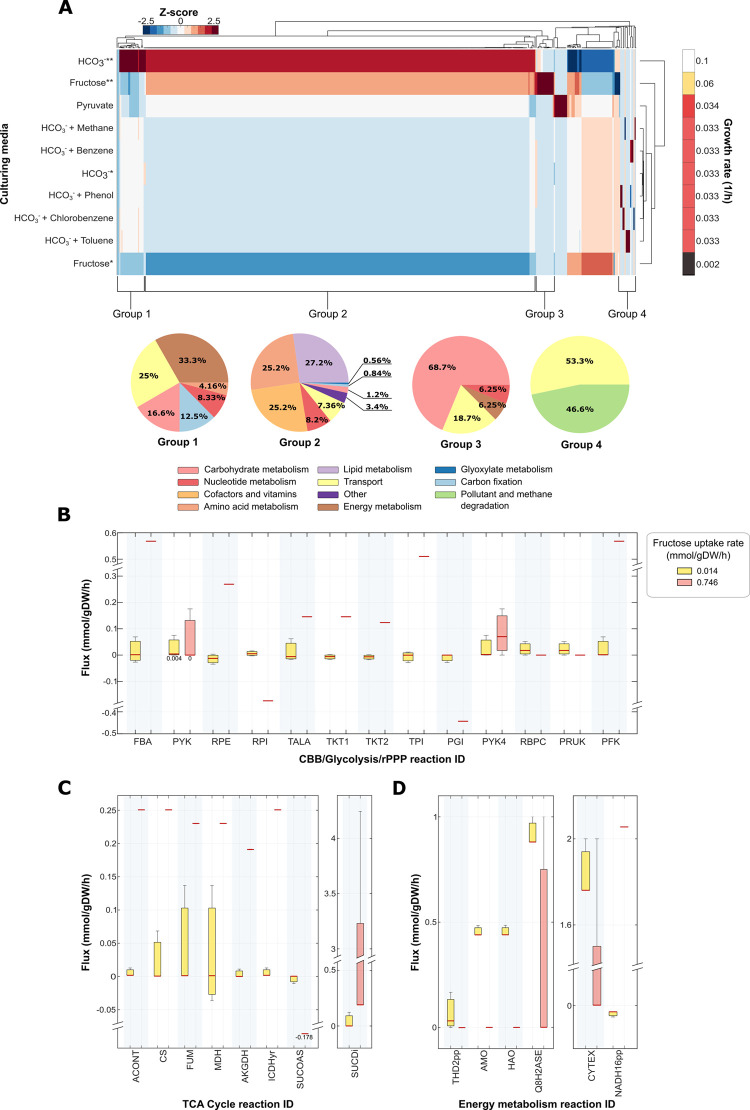
Fig 3. Flux distribution analysis under different growth conditions.
(A) Hierarchical clustering of flux distributions under different carbon sources and growth conditions. The HCO3- uptake rates were established at low and high levels, as we mentioned in Section 3.5.1. The pyruvate uptake flux (0.0773 mmol/gDW/h) used as a constraint in the simulations, resulted from the experimentally observed growth rate of 0.034 1/h [16] when Ne is grown using pyruvate as the organic carbon source. The methane and pollutant uptake rates were constrained to 1 mmol/gDW/h under the low HCO3- level condition. We used standararized Z-scores to normalize predicted fluxes. Z-scores represent negative and positive values in a blue to red color scale. The X-axis shows the metabolic reactions, and the Y-axis shows different growth conditions. * means low uptake rates; ** means high uptake rates. Group 1: 24 reactions; Group 2: 353 reactions; Group 3: 16 reactions; Group 4: 15 reactions. (B) Change in flux predictions of CBB cycle, glycolysis, rPPP reactions when fructose uptake is at low or high concentration. (C) Change in flux predictions of TCA cycle reactions when fructose uptake is at low or high concentration. (D) Change in flux predictions of energy metabolism reactions when fructose uptake is at low or high concentration. Abbreviations: FBA, Fructose-bisphosphate aldolase; PYK, pyruvate kinase; RPE, ribulose 5-phosphate 3-epimerase; RPI, ribose-5-phosphate isomerase; TALA, transaldolase; TKT, transketolase; TPI, triose-phosphate isomerase; PGI, glucose 6-phosphate isomerase; RBPC, ribulose 1,5-bisphosphate carboxylase-oxygenase; PRUK, phosphoribulokinase; PFK, phosphofructokinase; ACONT, aconitate hydratase; CS, citrate synthase; FUM, fumarase; MDH, malate dehydrogenase; AKGDH, 2-oxoglutarate dehydrogenase; ICDHyr, isocitrate dehydrogenase; SUCDi, succinate dehydrogenase; SUCOAS, succinyl-CoA synthetase; THD2pp, NAD(P)+transhydrogenase; AMO, ammonia monooxygenase; HAO, hydroxylamine oxidoreductase; Q8H2ASE, ubiquinol synthase; CYTEX, cytochrome exchange; NADH16pp, NADH dehydrogenase.