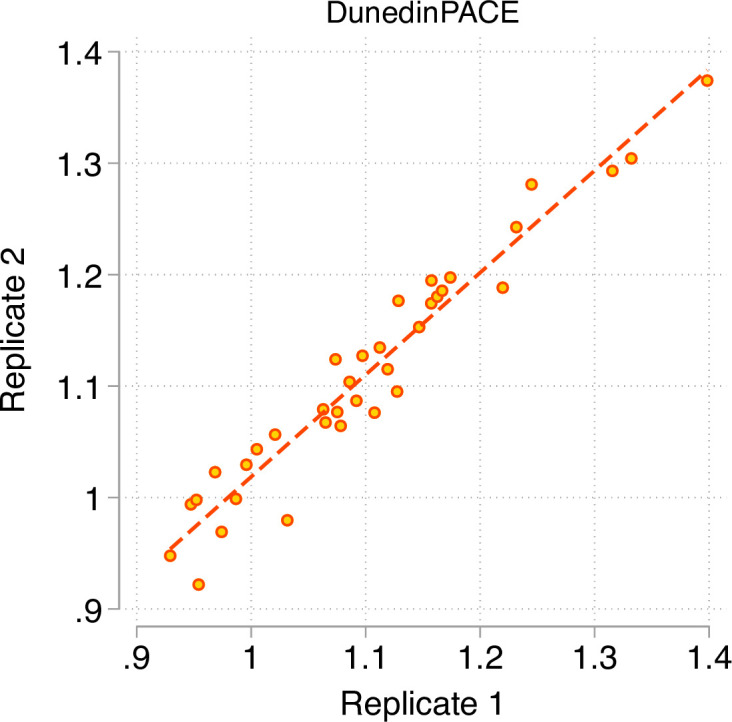
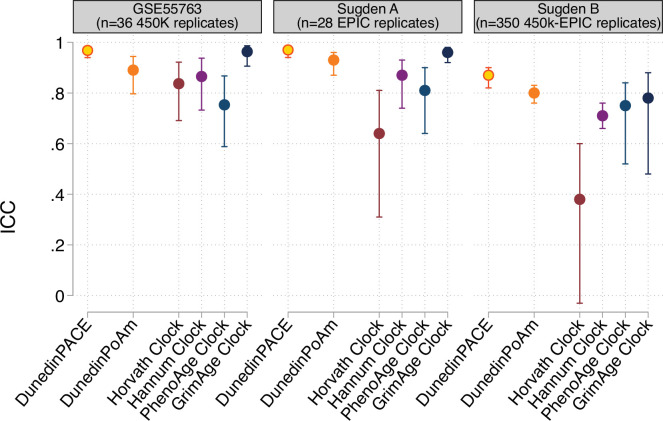
The figure plots intraclass correlation coefficients (ICCs) estimated from replicate DNA methylation datasets for DunedinPACE, original DunedinPoAm, and the DNA methylation clocks proposed by Horvath, Hannum et al., Levine et al. (PhenoAge), and Lu et al. (GrimAge). Error bars show 95% confidence intervals. Data are graphed from three datasets, 36 Illumina 450 k technical replicates from
GSE55763 (
Lehne et al., 2015), and 28 Illumina EPIC array technical replicates and 350 Illumina 450k-EPIC replicates from
Sugden et al., 2020. The Sugden et al. EPIC technical replicate dataset (Sugden A) included arrays analyzed as part of the Dunedin Study dataset. The Sugden et al. 450k-EPIC dataset (Sugden B) was used to identify probes for inclusion in the machine learning analysis from which DunedinPACE was derived.