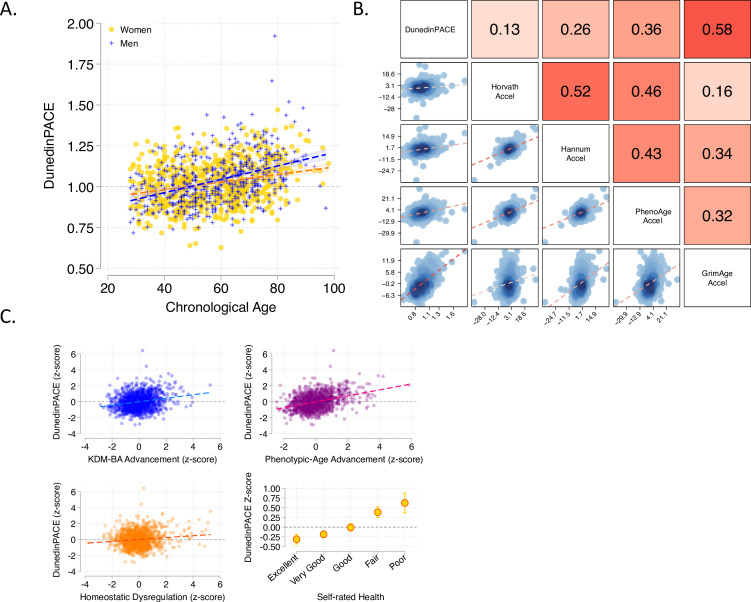
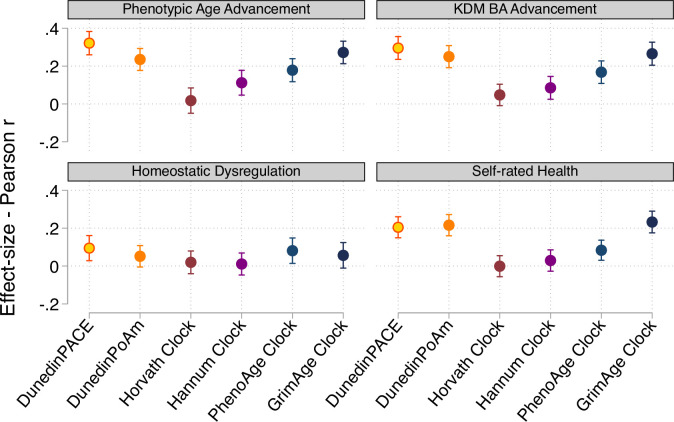
Figure 3. Associations of DunedinPACE with chronological age, epigenetic clocks, Physiology-based measures of biological age, and self-rated health in Understanding Society.
Panel A shows a scatterplot and fitted slopes illustrating the association between chronological age (x-axis) and DunedinPACE (y-axis) in women and men in the Understanding Society sample. Data for women are plotted with yellow dots (orange slope) and for men with blue crosses (navy slope). The figure illustrates a positive association between chronological age and DunedinPACE (Pearson r = 0.32). Panel B shows a matrix of correlations and association plots among DunedinPACE and age-acceleration residuals of Horvath, Hannum, Levine-PhenoAge and Lu-GrimAge epigenetic clocks. The diagonal cells of the matrix list the DNA methylation measures. The half of the matrix below the diagonal shows scatter plots of associations. For each scatter-plot cell, the y-axis corresponds to the variable named along the matrix diagonal to the right of the plot and the x-axis corresponds to the variable named along the matrix diagonal above the plot. The half of the matrix above the diagonal lists Pearson correlations between the DNA methylation measures. For each correlation cell, the value reflects the correlation of the variables named along the matrix diagonal to the left of the cell and below the cell. Panel C graphs scatterplots of associations of DunedinPACE with three physiology-based measures of biological age (KDM Biological Age Advancement, r = 0.30 [0.24–0.36]; Phenotypic Age Advancement, r = 0.32 95% CI [0.26–0.38]; and Homeostatic Dysregulation r = 0.09 [0.03–0.16]) and a plot of DunedinPACE means and 95% confidence intervals by self-rated health category (r = 0.20 [0.15–0.26]).