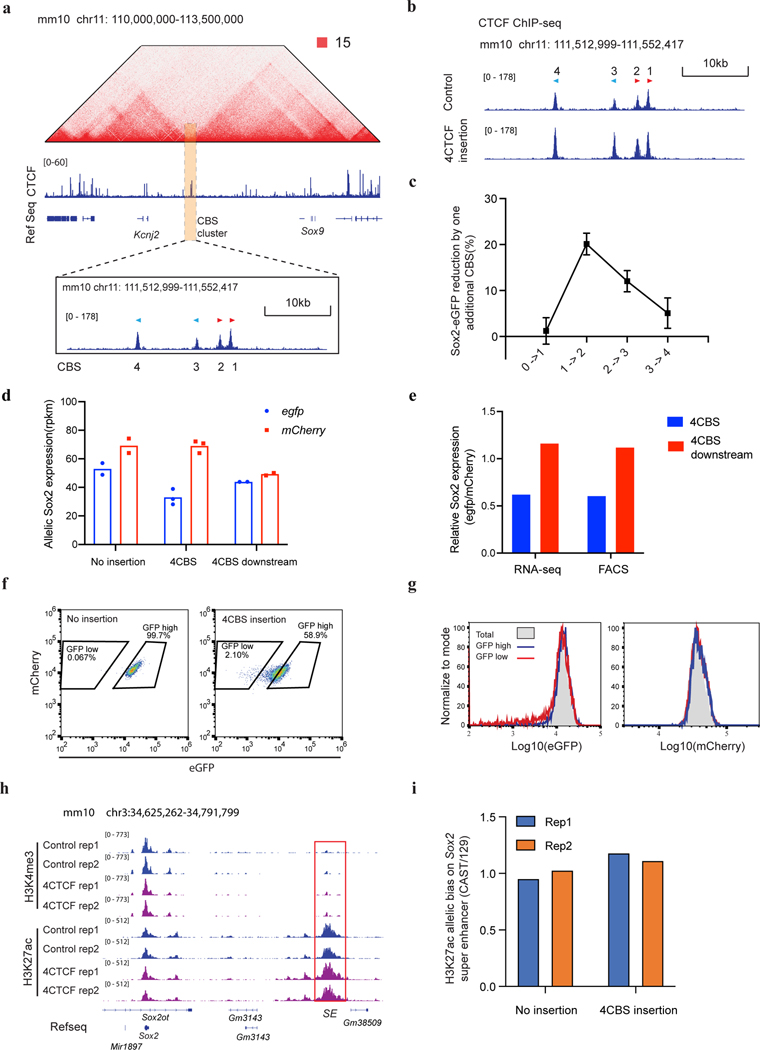
Extended Data Fig. 2. Insulation features of CBSs from the Sox9-Kcnj2 TAD boundary.
a, Hi-C contact map of the Sox9-Kcnj2 locus in mouse ES cells. Zoom in view shows the four CTCF binding sites cloned for insulator activity test. b, ChIP-seq of CTCF in the no insertion clone and the clone with an extra copy of the four Sox9-Kcnj2 TAD boundary CBS inserted inside the Sox2 domain. c, Reduction in Sox2-eGFP expression by one additional CBS. The comparison was between the clones presented in Figure 2b. (0 CBS, n = 8; 1 CBS inside, n = 23; 2 CBS inside, n = 18; 3 CBS inside, n = 13; 4 CBS inside, n = 5; Data are mean ± sd). d, Allele-specific Sox2 expression in the no insertion clone (n = 2), the 4CBS clone (n = 3) , and the 4CBS downstream clone (n = 2) as measured by RNA-seq. Sox2 expression from the CAST and 129 allele was represented by normalized read counts (rpkm) of the tagged egfp and mcherry gene, respectively. e, Relative Sox2 expression in the 4CBS and the 4CBS downstream clone in d measured by RNA-seq and FACS. The Sox2 expression from the egfp allele was first normalized to the mcherry allele, then compared to the no insertion clone. f, FACS profiling of the no insertion clone and the 4CBS clone. g, FACS profiling of GFPlow, GFPhigh sub-populations, and the unsort total population of the 4CBS insertion clone in f after extended culturing for 8 days. Left, GFP signal, right, mCherry signal from the same cells. h, ChIP-seq of H3K4me3 and H3K27ac in the no insertion clone and the 4CBS clone (n = 2). i, Allelic quantification of H3K27ac signal on the Sox2 super-enhancer of clones in h. H3K27ac ChIP-seq reads on the Sox2 super-enhancer were normalized by the total reads mapped to chromosome 3 for each allele.