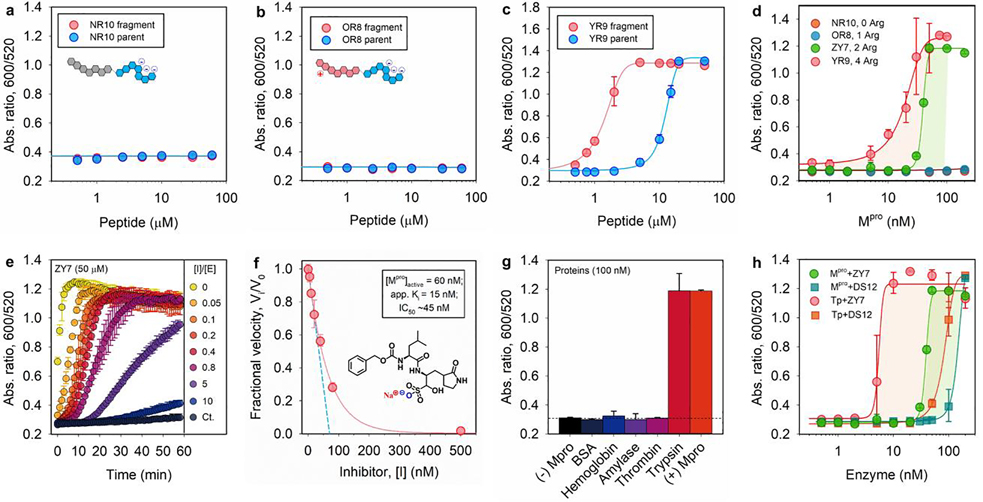
Figure 4.
Tunning sensitivity and specificity of the sensor. (a-c) The operation window of Mpro sensors based on NR10, OR8, and YR9 peptide, which contains 0, 1, and 4 arginine residues in its aggregating sequence, respectively; see Table 1. (d) The Mpro LoD of sensors based on four peptides of varying number of arginine: no LoD observed for NR10 and OR8 substrate; 27.7 and 3.4 nM is determined for YR9 and ZY7 substrate, respectively. (e) Time progression of ratiometric signal (Abs600/Abs520) in inhibitor assays. Increasing molar ratio of [inhibitor]/[Mpro] from 0−10 was employed. The control curve (Ct.) designates inhibitor only without Mpro additive. (f) A typical inhibition titration curve fitted with the Morrison equation (Eqn. S2) is shown for the competitive inhibitor, GC376.[30] Inset shows the chemical structure of GC376 inhibitor. A Henderson equation was applied to resolve the apparent inhibitor dissociation constant, Ki (app) =15 nM, and active enzyme concentration, [E]0 =60 nM (out of 100 nM). The IC50 is 45 nM. (g) Sensor activation by other mammalian proteins (100 nM), including bovine serum albumin (BSA), hemoglobin, α-amylase (100 U/mL), thrombin, and trypsin. Assay with and without Mpro is included as positive and negative control. (h) The response of sensors based on ZY7 and DS12 substrate to Mpro or trypsin in Tris buffer. LoD for Mpro is 27.7 and 114.4 nM for ZY7 and DS12, respectively; LoD for trypsin is 9.7 and 30.3 nM for ZY7 and DS12, respectively.