FIGURE 7.
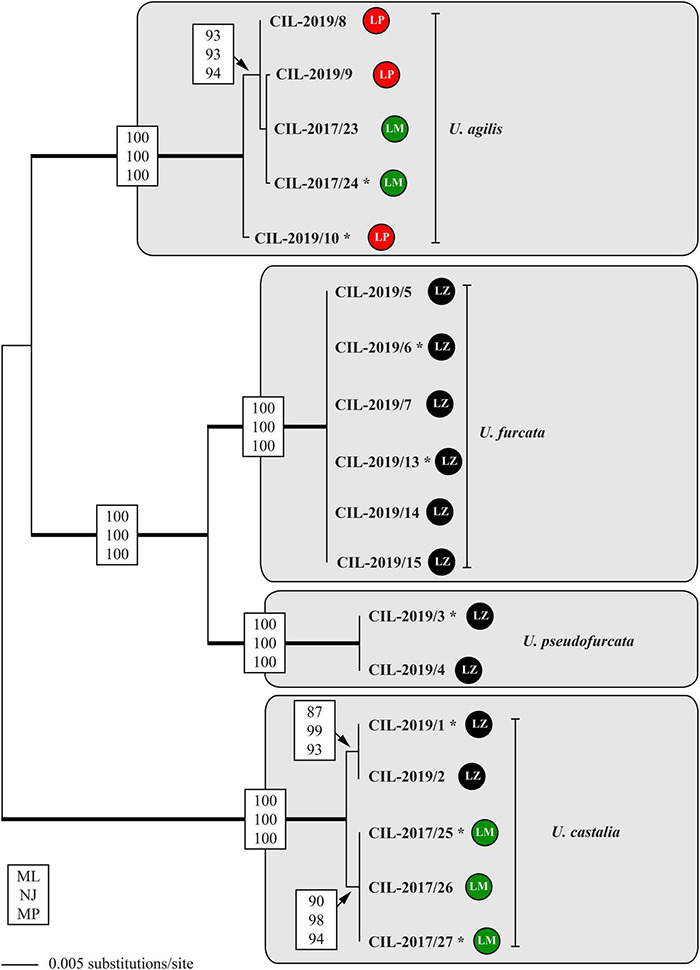
Molecular phylogeny of the genus Urotricha inferred from SSU and ITS rDNA sequences, using the maximum likelihood method of PAUP 4.0a167 on a dataset with 2,343 aligned positions of 18 taxa. For the analyses, the best model was calculated with automated model selection tool implemented in PAUP. The setting of the best model was given as follows: GTR + I (base frequencies: A 0.3017, C 0.1700, G 0.2153, T 0.3130; rate matrix A-C 1.3248, A-G 1.4840, A-U 1.8283, C-G 0.5727, C-U 2.8434, G-U 1.0000) with the proportion of invariable sites (I = 0.7703). The branches in bold are highly supported in all bootstrap analyses (bootstrap values > 50% calculated with PAUP, using the maximum likelihood, neighbor joining, and maximum parsimony). The circles following the strain designation indicate the strain origins (LP = Lake Piburg, Austria; LM = Lake Mondsee, Austria; LZ = Lake Zurich, Switzerland). Strains labeled with an asterisk were morphologically investigated.
