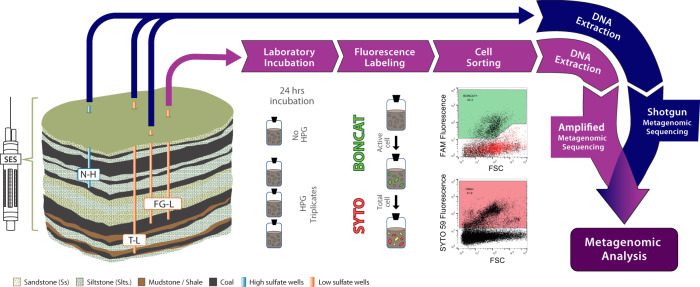
Fig. 1. Schematic depicting experimental workflow.
High-sulfate (Nance, N-L) and low-sulfate (Flowers Goodale, FG-L; Terret, T-L) CBM wells were sampled utilizing a Subsurface Environmental Sampler (SES) to retrieve in situ CBM samples of the attached fraction under environmentally relevant conditions. Following retrieval, samples were either prepared for BONCAT incubation (purple track) or shotgun metagenomics (blue track). For shotgun metagenomics, DNA was extracted and sequenced. BONCAT incubations with the synthetic amino acid tracer, HPG, lasted 24 h followed by cell removal from coal, click chemistry, staining, and cell sorting of BONCAT+ and BONCAT-Total (SYTO stained) cell fractions, DNA extraction and amplified metagenomic sequencing. The No HPG control was used to determine sorting gates for BONCAT+ events.