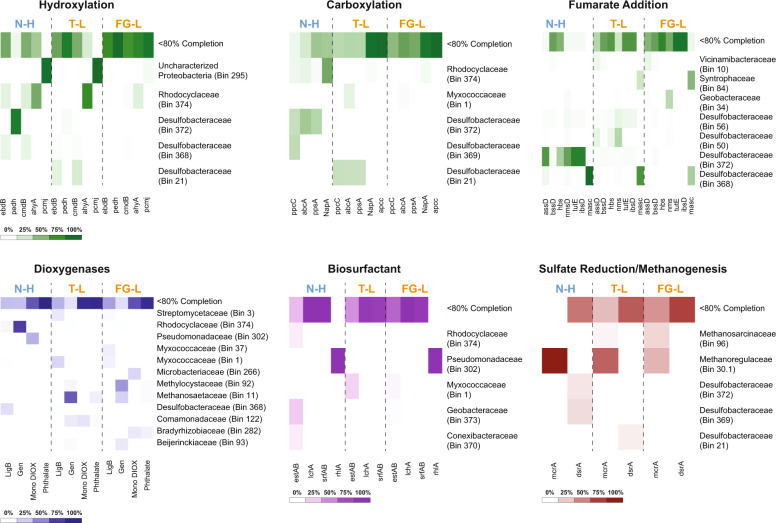
Fig. 3. Comparison of gene abundances across metagenome-assembled genomes MAGs.
Heatmaps from all MAGs from shotgun metagenomic samples depicting the relative gene abundance for individual genes involved in hydrocarbon degradation for the high-sulfate coal seam (N-H) and two low-sulfate coal seams (T-L and FG-L). Only high-quality MAGs (>80% completion) were taxonomically classified. All MAGS below 80% completion were grouped together as <80% Completion. The six different heatmaps, displaying the genes of interest, were grouped based on method of anaerobic degradation (green heatmaps) via hydroxylation, carboxylation, and fumarate addition, aerobic degradation (blue heatmap) via dioxygenases, biosurfactant production (purple heatmap), and sulfate reduction/ methanogenesis (red heatmap). All relative abundance of each gene found in the corresponding MAGs are represented by a color gradient (as seen in legend below heatmap) with the darkest color indicating the highest relative abundance for each sample and white indicating lack of the gene.