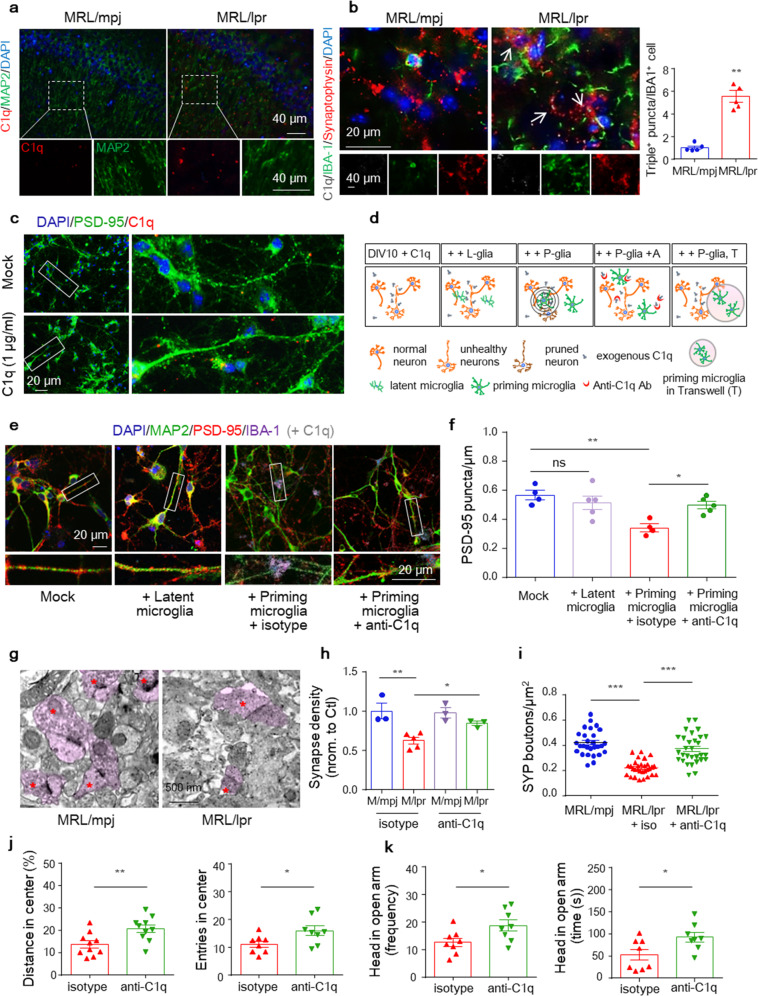
Fig. 5.
C1q tagging at synapses contributes to microglial synaptic elimination-mediated spine loss. a Immunostaining for C1q with the neuronal marker MAP2 in 6-week-old MRL/mpj and MRL/lpr mice. Scale bar, 40 μm. b Immunostaining showing colocalized synaptophysin, C1q, and IBA-1 puncta (with magnification insets and arrowheads depicting colocalization) in MRL/mpj and MRL/lpr mice. Scale bars as indicated. The data are the mean ± SEM. n = 5 mice per group, with an average of 3–4 slices per mouse. **P < 0.01 according to Student’s t-test. c MRL/lpr mouse-derived hippocampal neurons were cultured alone or with exogenous C1q and stained for PSD-95 (green), labeled C1q (red), and DAPI (blue). Scale bar, 20 μm. (d) Diagrams showing microglia–neuron (or transwell) cocultures and Sholl analyses to quantify synapses around microglia. Representative images of dendritic segments (e) and quantification of the dendritic PSD-95 puncta density (f) in neuron-microglia cocultures incubated with exogenous C1q and treated with anti-C1q antibodies or the isotype control. Primary neurons of MRL/lpr mice were cocultured with latent or primed microglia and stained for IBA-1 (purple), PSD-95 (red), and MAP2 (green). Scale bar, 20 μm. The data are the means of three independent experiments (f). *P < 0.05; **P < 0.01; and ns, not significant by one-way ANOVA with Tukey’s multiple comparisons test. g–i Representative transmission electron microscopy (TEM) images (g) and quantitation of synapse density (h) in hippocampal sections. Scale bars, 500 nm. (n = 3-5 mice per group). (i) Density of synaptophysin (SYP+) puncta in the CA3 region in control and anti-C1q-injected MRL/lpr mice. Each dot represents the value for one slice. *P < 0.05; **P < 0.01; and ***P < 0.001 according to one-way ANOVA with Tukey’s post hoc test. Parameters recorded and analyzed in the OFT (j) and EMP test (k) of MRL/lpr mice post antibody injection (n = 8–10 mice per group). *P < 0.05 and **P < 0.01 according to Student’s t-test. All data are the mean ± SEM. See also Supplementary Fig. 7