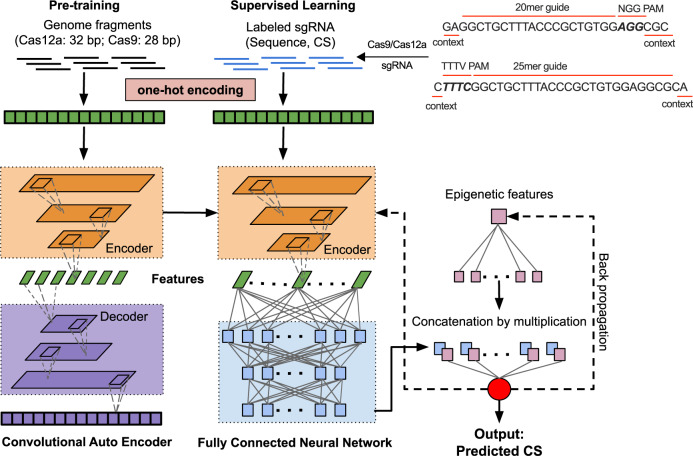
Fig. 3. The architecture of DeepGuide.
First, the entire Y. lipolytica PO1f genome was fragmented into sgRNA sized chunks (using a sliding window of 20 bp for Cas9 and 25 bp for Cas12a). Unsupervised pre-training was carried out on these unlabeled fragments using a convolutional autoencoder (left). The internal weights from the autoencoder were used to initialize a fully connected convolutional neural network (center). Labeled sgRNA (i.e., sequence and associated cutting score) were used as inputs for back-propagation learning on the fully connected neural network. See Supplementary Tables 2 and 3 for a description of the layers.