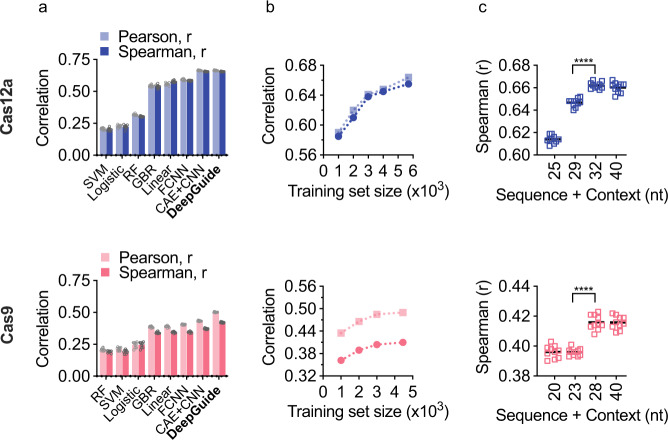
Fig. 4. Design and parameter optimization for DeepGuide on the Cas12a (top) and Cas9 (bottom) datasets.
a Evaluation of DeepGuide in a cross-validation analysis with several machine learning (ML) methods, including random forest (RF), support vector machines (SVM), logistic regression (Logistic), gradient boosting regression (GBR), linear regression (Linear), fully connected neural networks (FCNN), and the core architecture of DeepGuide, a combination of a convolutional autoencoder and a convolutional fully connected neural network (CAE + CNN). In addition to interconnected CAE and CNN, the final architecture of DeepGuide also includes a third fully connected network to account for nucleosome occupancy. Error bars indicate standard deviation over five independent cross-validation experiments. b The dependency of DeepGuide’s performance as a function of the training set size with smaller datasets produced by downsampling. c The dependency of DeepGuide’s performance as a function on the length of the context sequence around the sgRNA (tenfold cross-validation). One-way ANOVA indicates that sequence length has a significant effect (****p < 0.0001) for both Cas12a and Cas9. Tukey’s multiple comparison post hoc analysis indicates that for Cas12a the Spearman values for all sequence lengths, with exception of 32 vs. 40 bp (p = 0.708), are significantly different (p < 0.0001). For Cas9, Tukey’s multiple comparisons indicates that all values are significantly different (p < 0.0001) with the exceptions of 20 vs. 23 (p = 0.9995) and 28 vs. 40 bp (p > 0.9999).