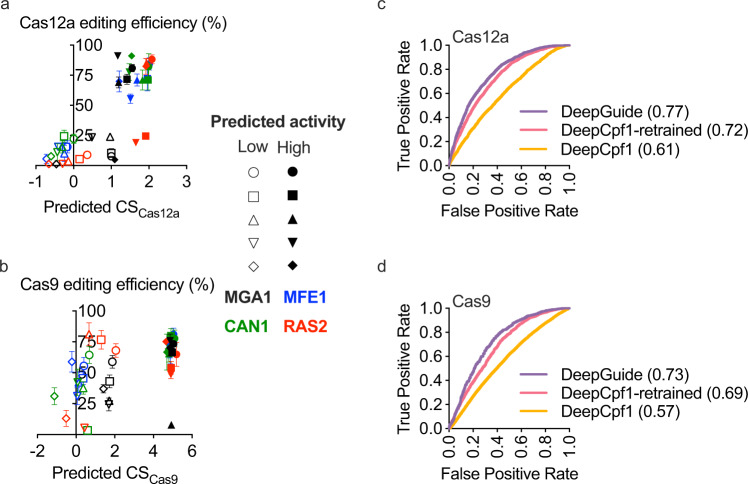
Fig. 5. External and internal validation of DeepGuide performance.
a, b Editing efficiencies of 5 predicted high-activity and 5 predicted low-activity sgRNA for Cas12a and Cas9 in single-gene disruption experiments. Genes MGA1, MFE1, CAN1, and RAS2 were picked as their null mutants displayed easily screenable phenotypes. Predicted high-activity sgRNAs clustered together, while low-activity sgRNAs clustered at lower editing efficiencies for Cas12a. Data points represent the mean of three biologically independent samples (n = 3), while the error bars represent the standard deviation. c, d ROC plots and AUROC values for DeepGuide prediction of high- and low-activity Cas9 and Cas12a sgRNAs.