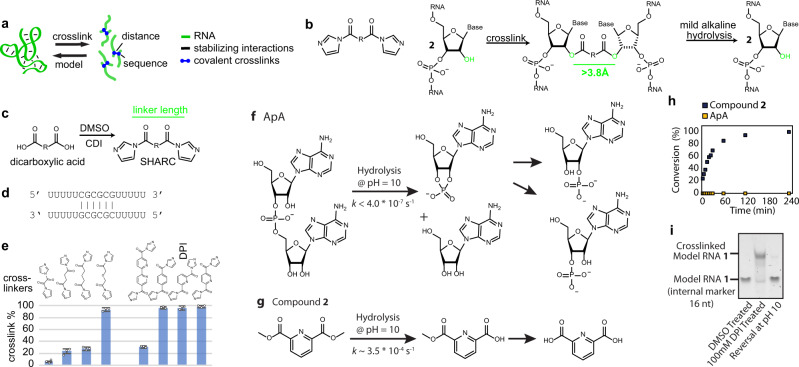
Fig. 1. Efficient RNA crosslinking and reversal using SHARC reagents.
a Principle of using crosslinking to determine RNA 3D structures. Spatial distances among nucleotides in an RNA should be sufficient to rebuild its 3D structure. b SHARC crosslinking and reversal of ribose 2′ hydroxyls. c One-step activation of dicarboxylic acids to produce SHARC reagents. CDI: carbonyldiimidazole. d Model RNA 1 homodimer duplex for testing SHARC crosslinking and reversal. e Crosslinking efficiency of a series of SHARC reagents on the model RNA 1 duplex. Common names for the 9 dicarboxylic acids to prepare the crosslinkers: oxalic, succinic, diglycolic, glutaric, 6,6′-binicotinic, terephthalic, dipicolinic, and isocinchomeronic. Crosslinking condition: MOPS buffer (pH 7.5 0.1 M KCl, 6 mM MgCl2), 4 h at room temperature. Data are mean ± s.d.; n = 3, technical replicates. f Hydrolysis kinetics of RNA phosphodiester bonds in a model dinucleotide ApA. g Hydrolysis of a model SHARC crosslinking product, compound 2. h Measurements of hydrolysis rates for the ApA and model compound 2 (5 mM starting concentration). i Example SHARC crosslinking and reversal of model RNA 1 duplex on a 20% urea-denatured TBE polyacrylamide gel. Source data are provided as a Source Data file.