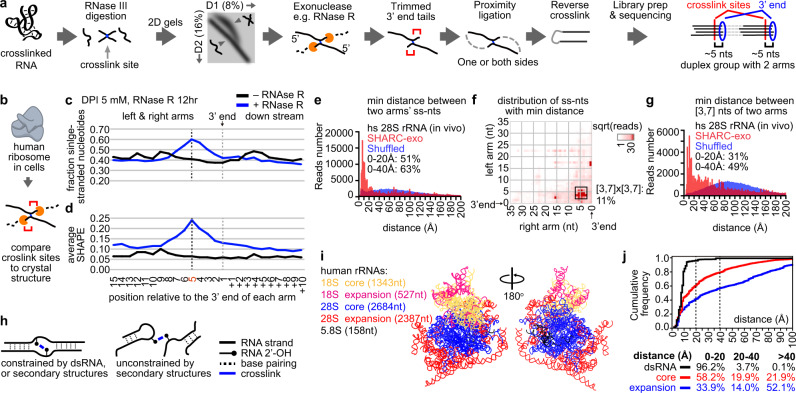
Fig. 2. Exonuclease trimming measures spatial distance at nucleotide resolution in cells.
a RNA samples, either in vitro or in cells, are crosslinked, digested to short fragments using RNase III, and the crosslinked fragments isolated using a DD2D gel. Isolated cross-linked fragments are then trimmed with exonucleases, proximally ligated, decrosslinked, ligated with adapters for cDNA library preparation. The putative crosslinking sites are determined based on the trimmed 3′ ends. b Benchmarking SHARC-exo against the human ribosome in cells. c, d Identification of SHARC crosslinking sites in the human ribosome from cross-linked cells. SHARC-exo condition: 5 mM DPI and 12 h RNase R trimming. Error bars represent standard deviations from two biological replicates. c Fraction of single-stranded nucleotides was calculated along each arm of the gapped reads. d Average icSHAPE signal was calculated along each arm of the gapped reads. e Distribution of minimal distances between single-stranded nucleotides (ss-nts) in the two arms of each gapped-read for the human 28S ribosomal RNA. The blue histogram is the distribution for randomly shuffled reads. f Positions of single-stranded nucleotides that are closest to each other on the two arms of each read. The square root of the reads was plotted. All reads were aligned relative to the 3′ end. Positions 3–7 for each arm were boxed, which represent 11% of total coverage. g Distribution of minimal distances between nucleotides in the 3–7 range on both arms. The blue histogram is the distribution for randomly shuffled reads. h Two types of SHARC crosslinked spatial proximity. i Illustration of the core and expansion segments (ESs) in three human cytoplasmic rRNAs. j Cumulative distribution of the minimal distances between the two arms of each read for reads mapped to different types of structures. Only non-dsRNA, or tertiary contact, reads are used for the core and expansion segments. Note, the color-coding in panels i and j have different meanings. Source data are provided as a Source Data file.