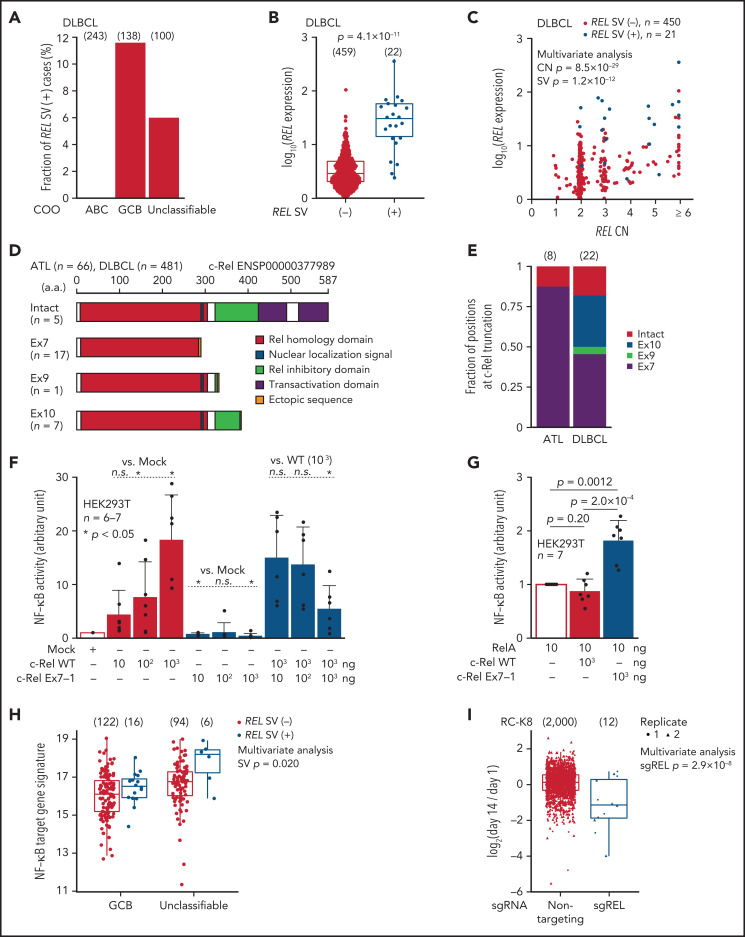
Figure 3.
REL-truncating SVs in DLBCL and its oncogenic function. (A) Frequency of aberrant REL transcripts in DLBCL according to cell-of-origin (COO). Numbers of cases are shown in parentheses. (B) Expression of REL exon 1-7 in 481 DLBCL cases from the National Cancer Institute Center for Cancer Research cohort35 analyzed by RNA-seq according to REL SV status. Numbers of cases are shown in parentheses. Two-sided Brunner-Munzel test. (C) Effect of REL CN and SV on REL exon 1-7 expression for DLBCL with available CN data (n = 471). Multivariate analysis using linear model. (D) Predicted structures of intact and representative truncated c-Rel proteins according to the truncation position. (E) Comparison of the truncation position of c-Rel proteins between ATL and DLBCL cases. Numbers of cases are shown in parentheses. (F-G) Luciferase assays of NF-κB transcriptional activity in HEK293T cells transduced to express WT and/or Ex7-1 c-Rel (F) and together with RelA (G) at indicated amount. (H) ssGSEA scores in GCB and unclassifiable DLBCL stratified by REL SV using a gene signature of NF-κB activation.46 Numbers of cases are shown in parentheses. Multivariate analysis using linear model. (I) Growth change of a REL SV-harboring DLBCL cell line (RC-K8) by clustered regularly interspaced short palindromic repeat (CRISPR)-mediated REL KO.36 Dot represents ratio of normalized abundance of each sgRNA in genome-scale CRISPR knockout library. Numbers of sgRNAs are shown in parentheses. Multivariate analysis using linear model. (B,H,I) Box plots show the medians (lines), IQRs (boxes), and ± 1.5 × IQR (whiskers). (F-G) Data represent means + standard deviation. Two-sided Welch's t test. Ex, exon; n.s., not significant.