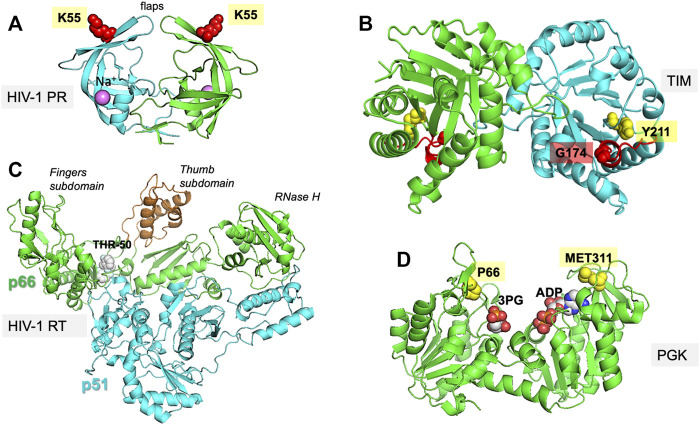
FIGURE 1.
Proteins investigated in the present study. The figure displays the experimentally resolved X-ray structures also used as initial structures for simulations. (A) HIV-1 protease (PR) (PDB id: 1tw7) is a wide-open, apo structure. The residue K55 on each subunit of the homodimer isused to probe the opening/closure of the flaps. (B) TIM (PDB id:1tcd) is a homodimeric enzyme, for which the catalytic loop is shown in red on both subunits of the apo state. The distance between the catalytic loop tip residue G174 and Y211 defines loop opening/closure motion in each subunit. (C) HIV-1 RT (PDB id: 2b6a) is a heterodimer composed of p51 and p66 subunits. The current structure is in complex with THR-50. The distance between the fingers and thumb subdomains, both located on the p66 subunit, indicate a transition between closed and open conformations of the region between these two subdomains. (D) PGK (PDB id: 2xe7) in the presence of the two substrates, 1,3-bisphosphoglycerate (bPG) and ADP. The distance between P66 and M311, two residues located at the tips of the N- and C-domains near the ligands, probes the opening/closing movement of the enzyme required for its catalytic activity.