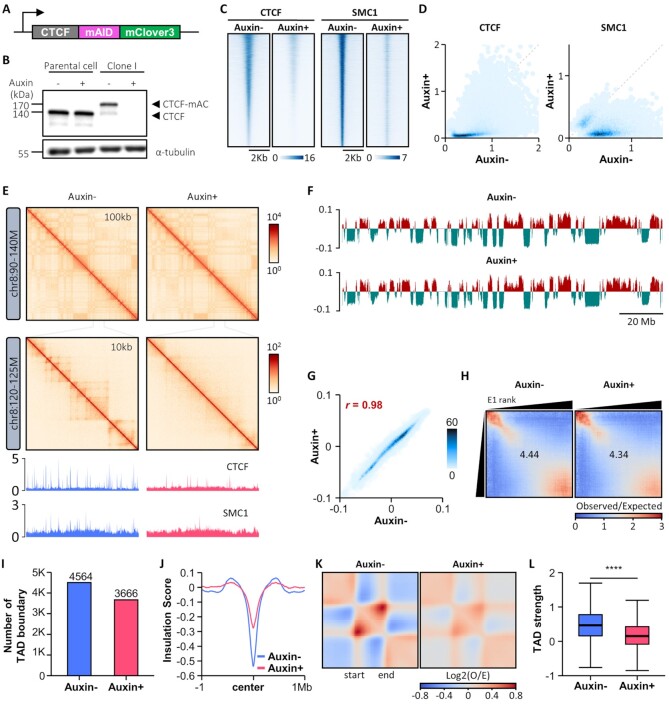
Figure 1.
Acute depletion of endogenous CTCF protein maintains compartment organization, but compromises global TAD insulation. (A) The mAID-mClover3 cassette for auxin-inducible degron is tagged to the C-terminus of CTCF on both alleles in a HCT116 cell line wherein OsTIR1 is integrated at the AAVS1 locus. (B) Immunoblotting shows complete degradation of endogenous mAID-mClover3-tagged CTCF (CTCF-mAC) upon auxin treatment. (C and D) Heatmaps (C) and scatterplots (D) of ChIP-Seq signals called for CTCF (left) and SMC1 (right) show marked reductions in the global occupancies of both proteins upon auxin treatment. (E) Hi-C contact maps generated by HiCExplorer at 100- and 10-kb resolution (version 3.4.3) (113). ChIP-seq signal tracks for CTCF and SMC1 were aligned below the contact maps at 10-kb resolution. (F) Distributions of cis Eigenvector 1 values across the entirety of chromosome 8 with and without auxin treatment. (G) Scatterplot shows that CTCF depletion does not affect genome-wide cis Eigenvector 1 values. (H) Saddle plots of compartmentalization strength with and without auxin treatment. (I) Number of TAD boundaries obtained with Hi-C data. (J) Genome-wide averaged insulation scores plotted against distance around insulation center at WT TAD boundaries. (K) Heatmaps show the average observed/expected Hi-C interactions in the TAD regions. (L) Boxplot shows TAD strength with and without auxin treatment.