Fig. 1 |. HR is stimulated by local transcription.
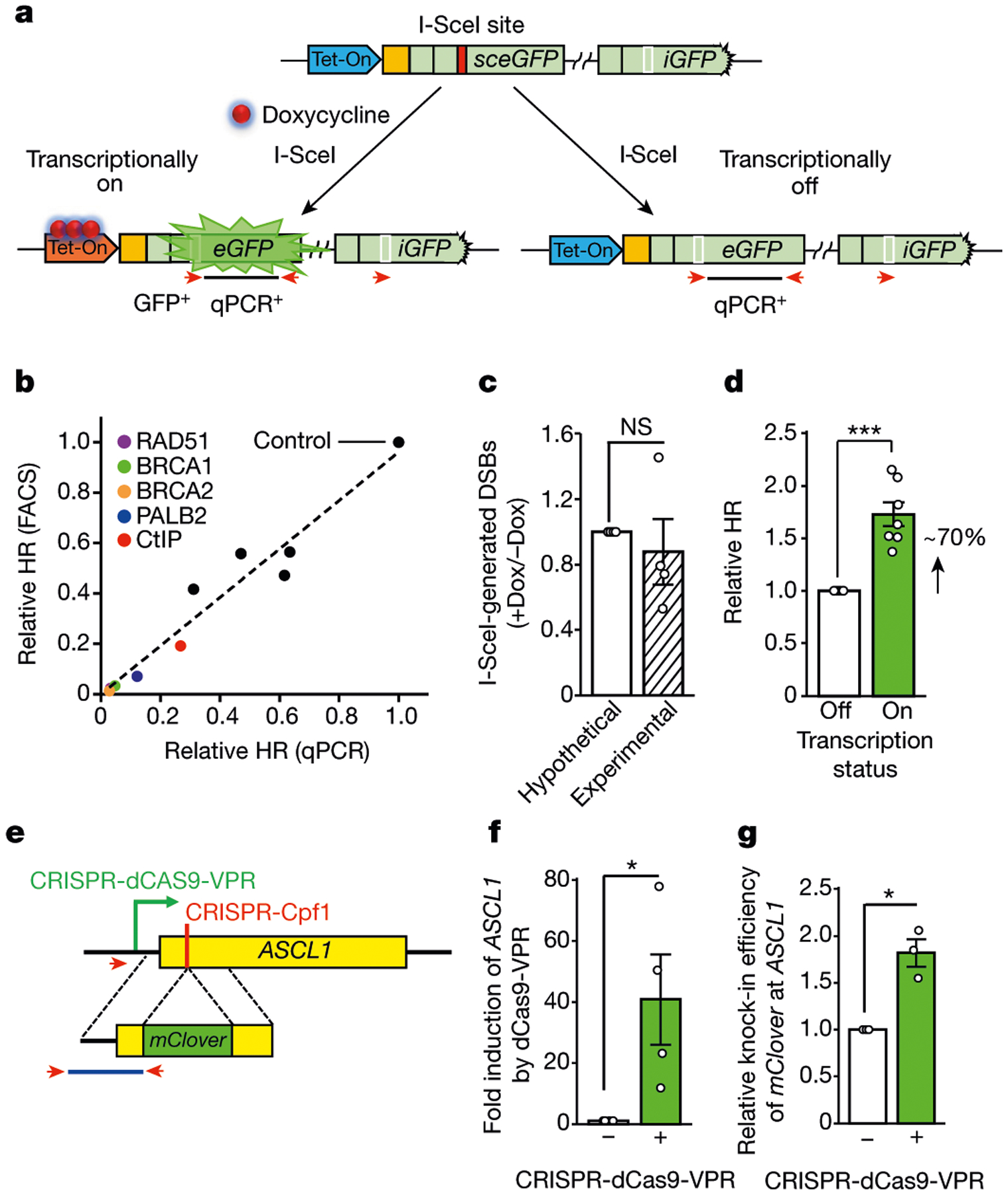
a, Schematic of the Tet-DR-GFP reporter. The coding and 5′ untranslated regions of GFP are in light green. A unique sequence upstream of sceGFP is in orange. A Tet-On promotor (blue) is upstream of sceGFP. After the I-SceI site (red box) is repaired, the wild-type eGFP sequence (white box) is restored. The repair product is detected by qPCR using specific primers (red arrows). b, Spearman correlation between qPCR- and fluorescence-activated cell sorting (FACS)-based HR assays (R = 0.99, P < 0.0001). Each dot represents a cell population depleted of an HR factor (Extended Data Fig. 1c). c, DNA fragments containing I-SceI-generated DNA ends were captured with biotinylated complementary oligos and quantified by qPCR. The ratio of DSB levels in samples without doxycycline (−Dox) and with doxycycline (+Dox) was determined. Data are mean ± s.e.m. (n = 4 independent experiments). NS, not significant (P ≥ 0.05) (two-sided Student’s t test; P = 0.59). d, HR efficiency was measured by qPCR in transcriptionally on and off states (+Dox and −Dox, respectively). The HR efficiency in −Dox is 1. Data are mean ± s.e.m. (n = 7 independent experiments). ***P <0.001 (two-sided Student’s t test; P = 0.0007). e, Schematic of the HR assay using the ASCL1 gene. CRISPR-dCas9-VPR activates ASCL1 transcription. CRISPR-Cpf1 generates a DSB near the 5′ end of the ASCL1 coding region (yellow). The homologous sequences flanking mClover are bracketed by dotted lines. The knock-in of mClover to ASCL1 was quantified by qPCR using specific primers (red arrowheads). f, Transcriptional activation of ASCL1 by CRISPR-dCas9-VPR. The level of ASCL1 mRNA in the absence of CRISPR-dCas9-VPR is 1. Data are mean ± s.e.m. (n = 4 independent experiments). *P < 0.05 (one-sided Student’s t test; P = 0.037). g, Relative knock-in efficiency of mClover at ASCL1 with or without CRISPR-dCas9-VPR. Data are mean ± s.e.m. (n = 3 independent experiments). *P < 0.05 (two-sided Student’s t test; P = 0.031).
