Extended Data Fig. 10 |. Characterizations of RNA transcripts and DNA–RNA hybrids at sceGFP and iGFP loci.
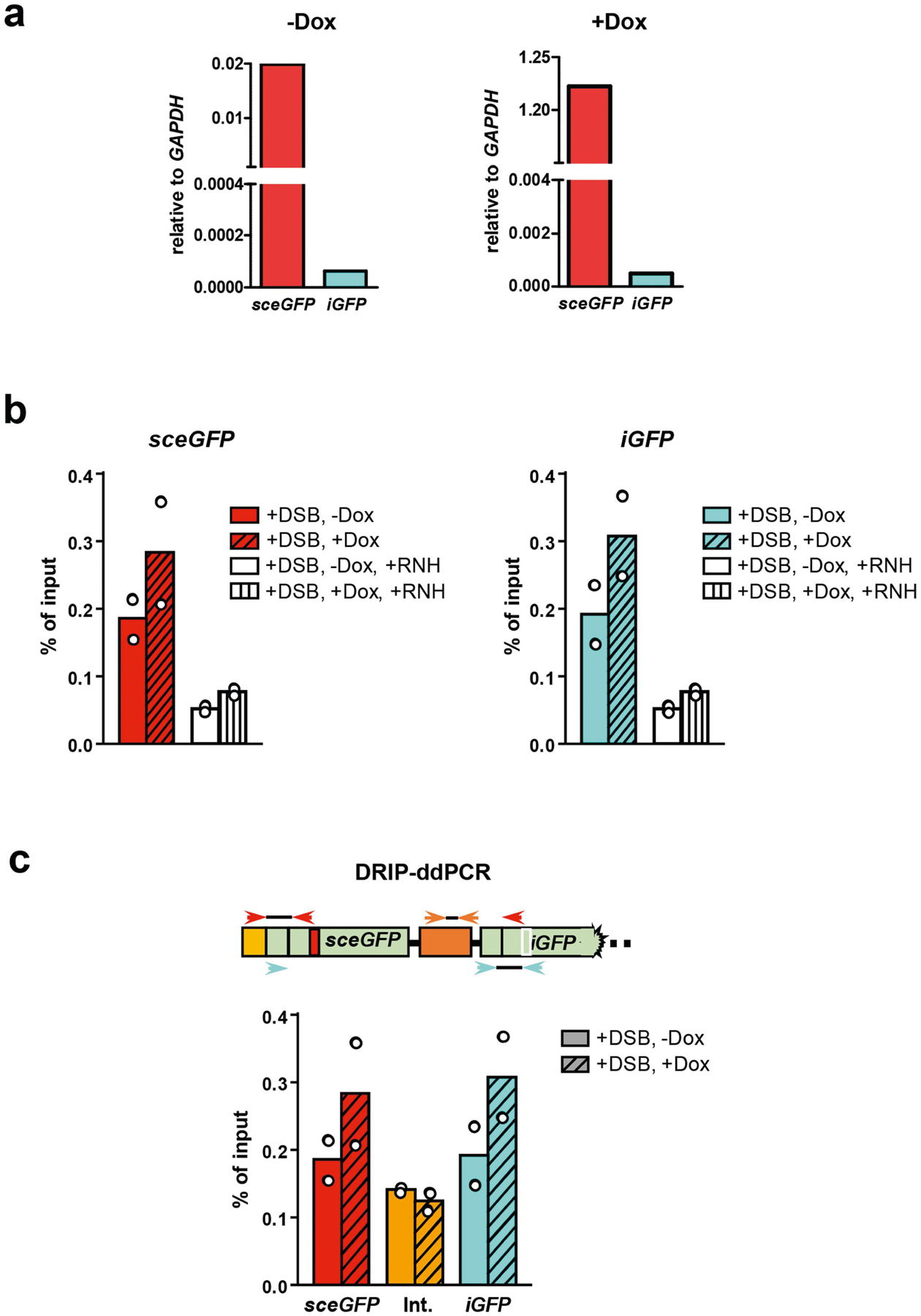
a, Total RNA isolated from U2OS-Tet-DR-GFP cells treated or untreated with Dox were digested with dsDNase and reverse-transcribed using random primers. cDNA was subjected to qPCR analysis to determine the relative levels of sceGFP and iGFP transcripts using the GAPDH transcript as a reference. The sceGFP transcript was increased by more than 60-fold by Dox. In the absence of Dox, the sceGFP transcript was more than 300-fold more abundant than the iGFP transcript. In the presence of Dox, the sceGFP transcript was more than 2,500-fold more abundant than the iGFP transcript. These results suggest that only sceGFP, and not iGFP, is transcribed in the presence of Dox. b, The accumulation of DNA–RNA hybrids in sceGFP and iGFP was analysed by DRIP–ddPCR as in Fig. 4f. In the +RNH (RNaseH) samples, extracted total nucleic acids were treated with RNaseH before being subjected to DRIP analysis. Data are mean (n = 2 independent experiments). c, The levels of DNA–RNA hybrids were tested by DRIP–ddPCR using primers that specifically detect an internal region between sceGFP and iGFP (orange arrowheads). These specific primers do not detect any sceGFP or iGFP containing DNA fragments after the restriction digestion during DRIP–ddPCR. The levels of DNA–RNA hybrids were measured in transcriptionally on and off states (+/− Dox) after DSB induction (+I-SceI). Data are mean (n = 2 independent experiments).
