Fig. 4 |. RAD51AP1 drives R-loop formation in vitro and in cells.
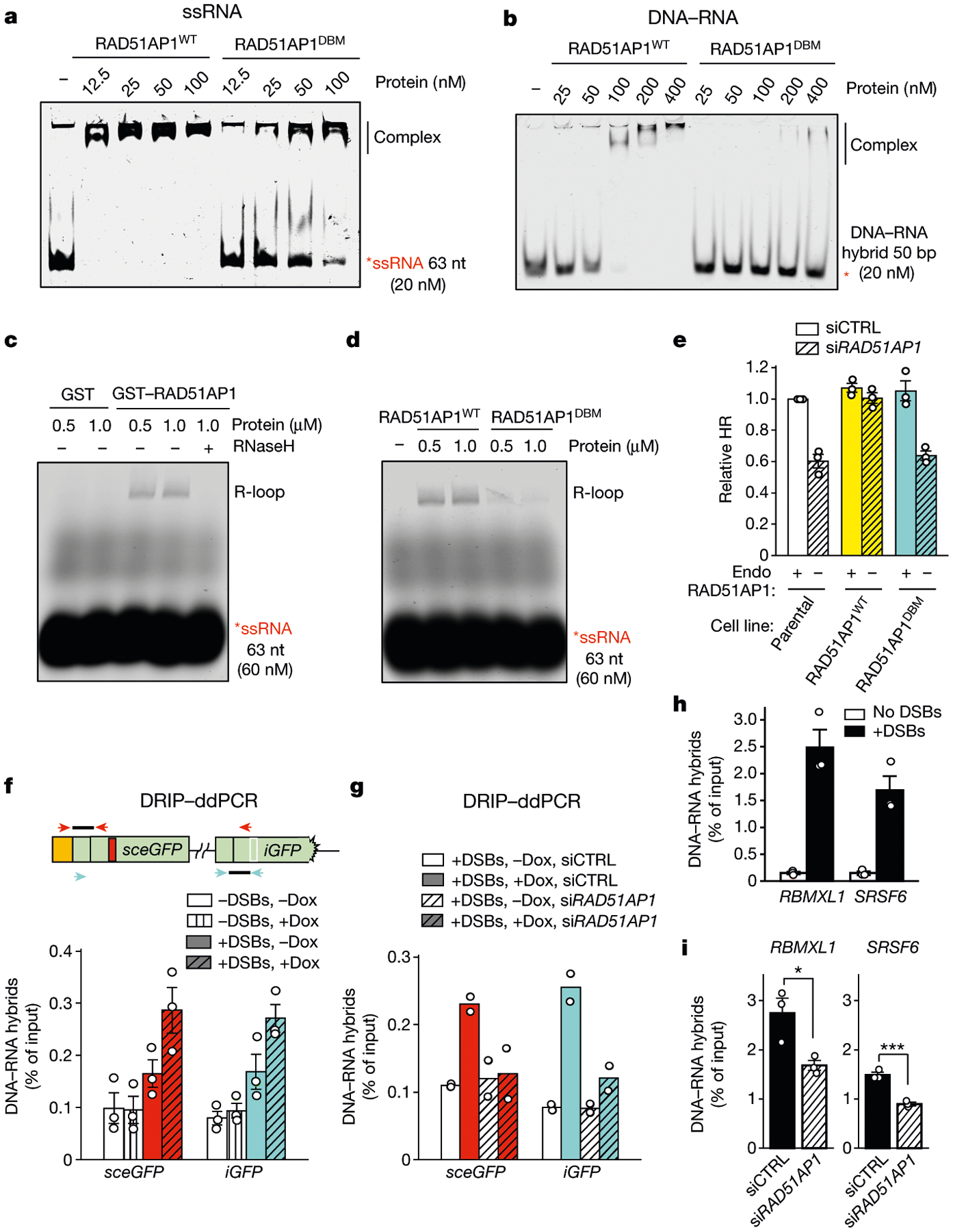
a, b, Increasing concentrations of RAD51AP1WT or RAD51AP1DBM were incubated with labelled ssRNA (a) or DNA–RNA hybrid (b). The formation of complexes was analysed by EMSA. c, d, Increasing concentrations of GST and GST–RAD51AP1 (c), or RAD51AP1WT and RAD51AP1DBM (d), were incubated with labelled ssRNA and a dsDNA plasmid. The formation of R-loops was analysed with agarose gel. Treatment with RNaseH (2.5 U, 5 min) eliminated R-loops (c). e, HR efficiency was measured by DR-GFP assay in cells stably expressing siRNA-resistant RAD51AP1WT or RAD51AP1DBM and transfected with control siRNA (siCTRL) or siRNA targeting RAD51AP1 (siRAD51AP1). Endo, endogenous. Data are mean ± s.e.m. (n = 3 independent experiments). f, The levels of DNA–RNA hybrids in sceGFP and iGFP were analysed by DRIP–ddPCR using primers specific for sceGFP (red arrowheads) or iGFP (cyan arrowheads). Hybrids were measured in transcriptionally on and off states (+Dox and −Dox, respectively) and with or without I-Sce1-induced DSBs. Data are mean ± s.e.m. (n = 3 independent experiments). g, Hybrids in sceGFP and iGFP were analysed as in f in control and RAD51AP1-knockdown cells. Data are mean (n = 2 independent experiments). h, i, Hybrids at two AsiSI sites in active genes were measured in the presence or absence of DSBs (h), or in control and RAD51AP1-knockdown cells (i). Data are mean ± s.e.m. (n = 3 independent experiments). *P < 0.05; ***P < 0.001 (two-sided Student’s t test; P = 0.029 for RBMXL1; P = 0.0008 for SRSF6).
