Extended Data Fig. 1 |. Characterizations of Tet-DR-GFP and ASCL1-mClover HR reporters.
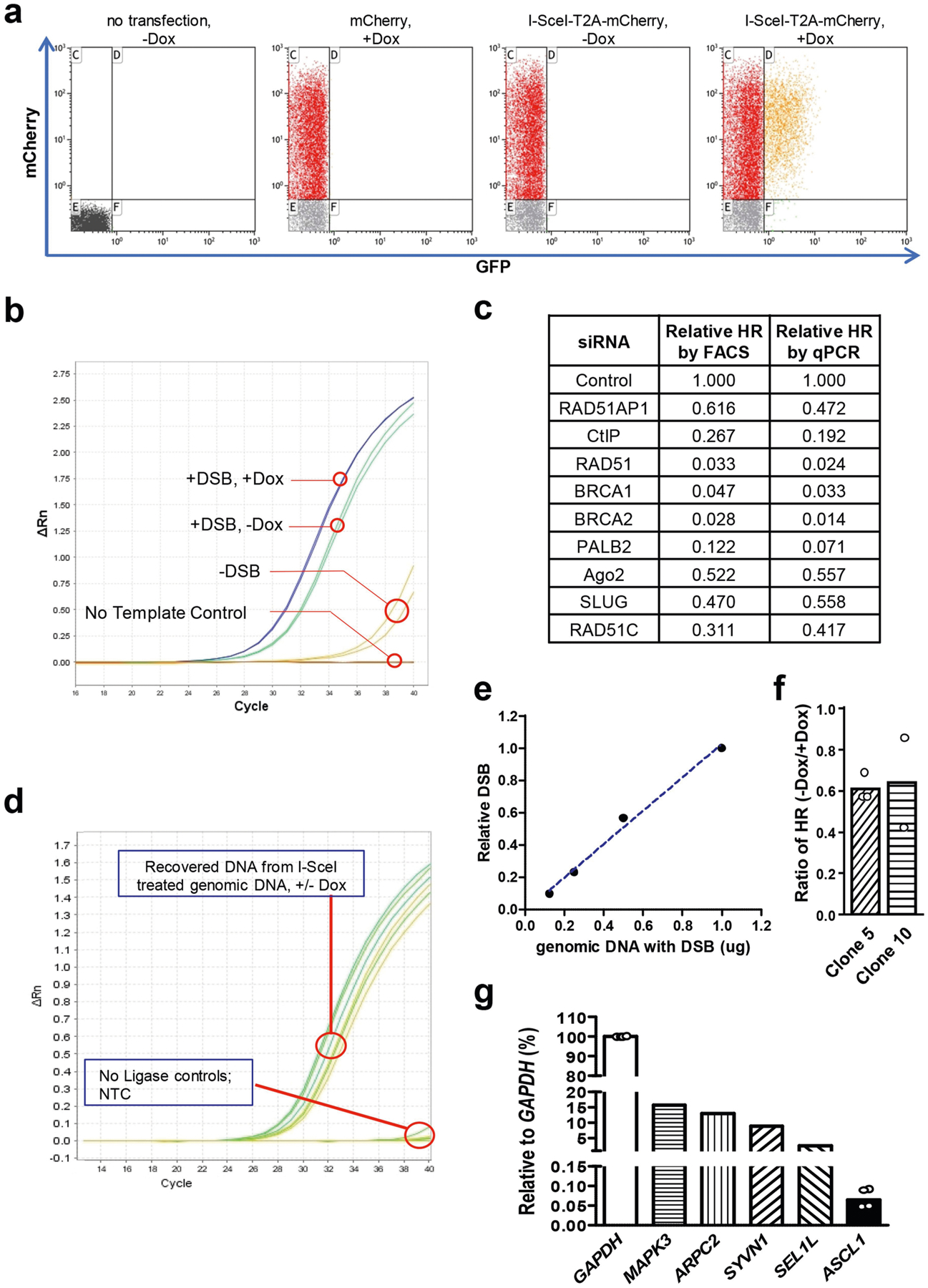
a, Flow cytometry analysis of a U2OS-derivative cell line in which the Tet-DR-GFP reporter is stably integrated. Cells were mock transfected, transfected with a control plasmid expressing mCherry or transfected with a plasmid expressing I-SceI-T2A-mCherry. GFP+ cells were only detected when I-SceI-T2A-mCherry was expressed and transcription of sceGFP was induced by Dox. b, Detection of repaired eGFP by qPCR. Cells carrying the Tet-DR-GFP reporter were mock transfected or transfected with a plasmid expressing I-Sce-T2A-mCherry (−/+ DSBs) in the absence and presence of Dox (−/+Dox). Genomic DNA was analysed for the repaired eGFP sequence by qPCR. c, FACS- and qPCR-based HR assays detect changes of HR efficiency similarly. Cells were transfected with siRNAs to knock down the indicated HR proteins and analysed by FACS- and qPCR-based HR assays. d, e, The transcription status of sceGFP does not affect I-SceI induced DSB formation. The I-SceI-induced DNA ends in sceGFP were ligated to a sequence-specific adaptor and quantified by qPCR. d, Amplification plot of qPCR. e, Linear quantification of DSBs by qPCR. The indicated amounts of genomic DNA containing I-SceI-generated DSBs were mixed with uncut genomic DNA, so that a total of 1 μg genomic DNA was analysed in each qPCR sample. f, Two additional stable U2OS clones carrying the Tet-DR-GFP reporter were used to analyse the ratio of HR levels in transcriptionally off and on states (−Dox/+Dox). Data are mean (n = 2 independent experiments). g, Total RNA isolated from HEK293T cells was reversely transcribed using random primers. cDNA was subjected to qPCR analysis to determine the relative expression levels of CRISPR-dCas9-VPR-activated ASCL1 and a panel of endogenous genes using the GAPDH transcript as a reference. Data of ASCL1 are mean (n = 4 independent experiments).
