Extended Data Fig. 3 |. Effects of depletion of RAD51AP1, UAF1 and RAD52 on HR.
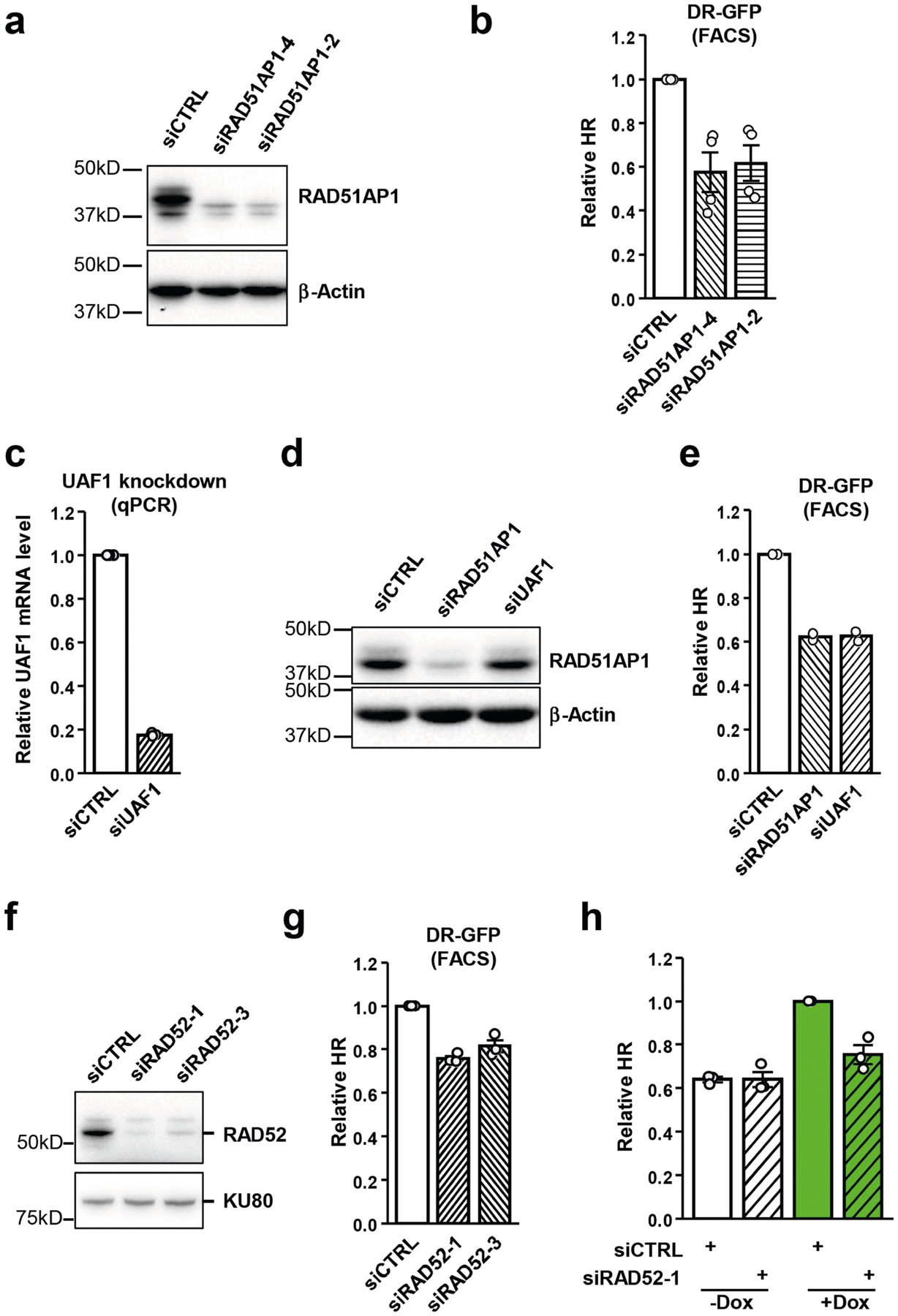
a, Cells were transfected with control or two independent RAD51AP1 siRNAs. Levels of endogenous RAD51AP1 and β-actin (loading control) were analysed by western blot. A representative western blot of three similar experiments is shown. b, HR efficiency was measured by FACS using U2OS-Tet-DR-GFP reporter cells after knockdown of RAD51AP1. Data are mean ± s.e.m (n = 3 independent experiments). c, Cells were transfected with control or UAF1 siRNA. Levels of UAF1 mRNA were analysed by RT–qPCR. Data are mean (n = 2 independent experiments). d, Cells were transfected with control, RAD51AP1 or UAF1 siRNA. Levels of endogenous RAD51AP1 and β-actin (loading control) were analysed by western blot. A representative western blot of three similar experiments is shown. e, HR efficiency was measured by FACS using U2OS-Tet-DR-GFP reporter cells after knockdown of RAD51AP1 or UAF1. Data are mean (n = 2 independent experiments). f, Cells were transfected with control or two independent RAD52 siRNAs. Levels of endogenous RAD52 and KU80 (loading control) were analysed by western blot. A representative western blot of three similar experiments is shown. g, HR efficiency was measured by FACS using U2OS-Tet-DR-GFP reporter cells after knockdown of RAD52. Data are mean ± s.e.m (n = 3 independent experiments). h, HR efficiency was measured by qPCR in transcriptionally on and off states (+/−Dox) after knockdown of RAD52. The HR efficiency of control siRNA transfected cells in the transcriptionally on state (+Dox) serves as a reference. Data are mean ± s.e.m (n = 3 independent experiments).
