Fig. 2. PAX7 expression timing in quail, chick, and peafowl whole embryos.
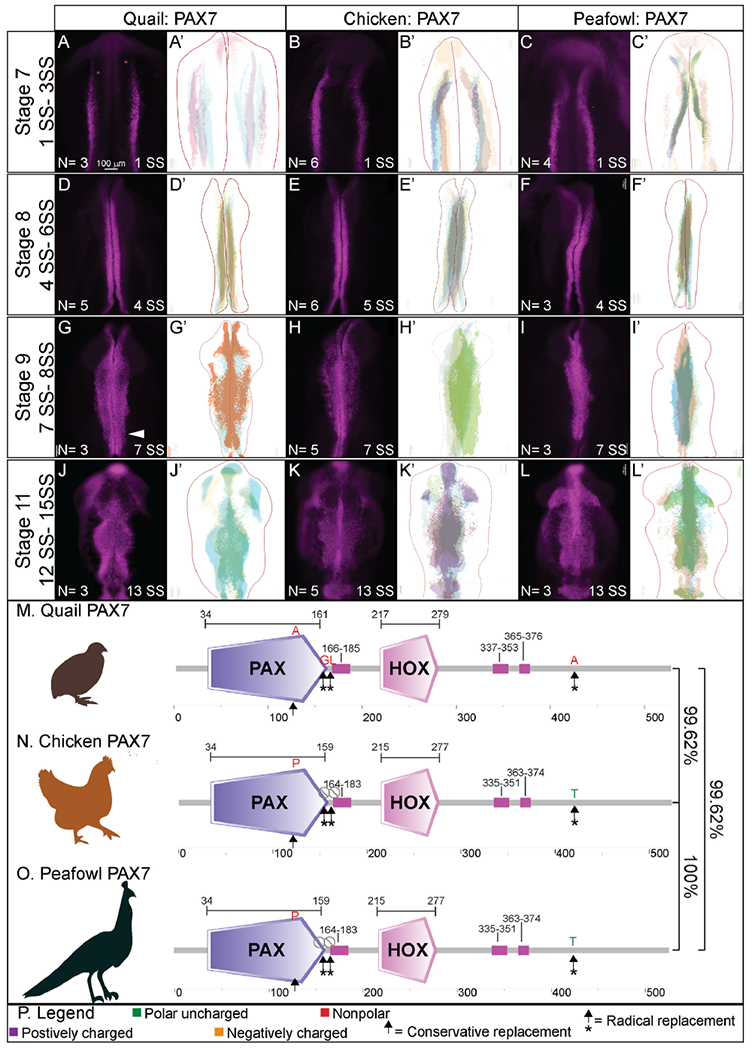
IHC for PAX7 expression in (A-C′) HH7 (1 SS) neural plate border, (D-F′) HH8 (5 SS) dorsal neural tube, (G-I′) HH9 (7 SS) EMT stage NC cells, and (J-L′) HH11 (13 SS), neural tube and migratory NC cells. (A, D, G, J) in quail, (B, E, H, K) in chick, and (C, F, I, L) in peafowl embryos. (A′, B′, C′) schematic overlays of multiple embryos. Quail exhibits more posterior midline PAX7+ expression at HH9 (G, arrowhead). Number of embryos analyzed at each stage and used for schematic overlays indicated in IHC panels. Scale bar is 100 μm and all images were taken at the same magnification. (M–O) Amino acid sequences were aligned and compared, then analyzed in SMART to obtain domain diagrams. PAX (purple) and HOX (pink) domains are indicated on images and small pink boxes are low complexity domains. (N, O) Chick and peafowl PAX7 proteins are identical and (M) quail PAX7 has two amino acid replacements, one conservative (same type of amino acid, black arrow), one radical replacement (black arrow with asterisk), and two additional amino acids (black arrows with asterisks). (P) Legend for (M–O). Purple is positively charged amino acid, green is polar uncharged, yellow is negatively charged, red is nonpolar. Scale bar for all whole mount images is 100 μm and is marked in the first image.
