Fig. 14.
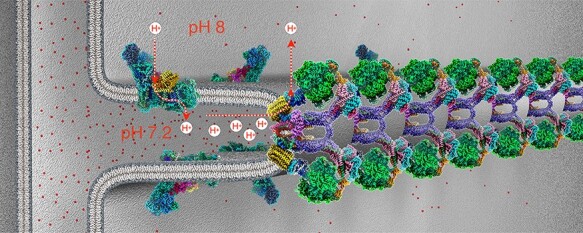
Molecular organization of mitochondrial inner membrane cristae. Cryo-EM structures of the F1Fo ATP synthase dimers [58] and complex I [120], both from Polytomella sp. in the membrane. Three-dimensional density maps at 2.7 Å (ATP synthase) and 2.9 Å resolution (complex I) are coloured by subunit. ATP synthase dimers assemble into long rows that impose a tight local curvature on the inner membrane, giving rise to the formation of cristae as mitochondrial microcompartments. Complex I and other respiratory chain complexes (not shown) occupy the flat membrane regions on either side of the dimer rows. Complex I generates most of the proton gradient across the inner membrane, pumping protons (dashed red arrows) into the cristae, which work as proton traps. Protons (red dots) flow through channels in the Fo subcomplex of the ATP synthase from the cristae lumen (∼pH 7.2) to the mitochondrial matrix (pH 8), driving the rotor (yellow) in the membrane. The torque generated by the electrochemical gradient is transmitted via the central stalk (blue) to the catalytic F1 subcomplex (green), where ATP is produced from ADP and phosphate by rotary catalysis.
