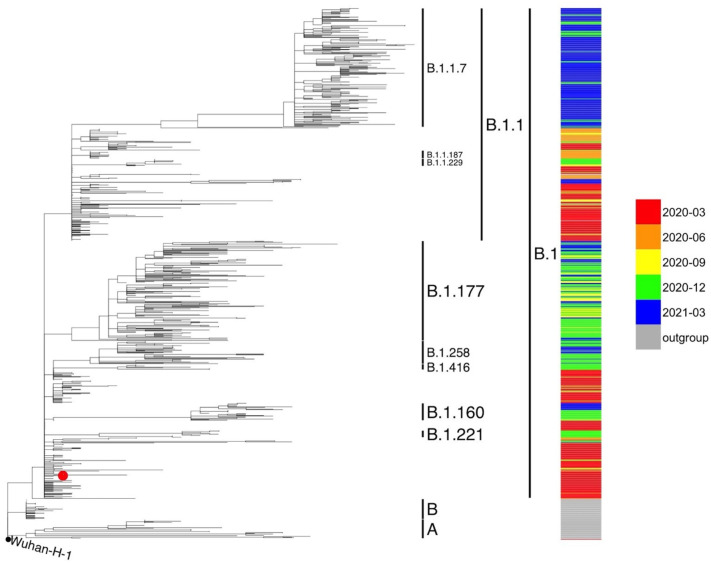
Fig. 1.
Phylogenetic analysis of SARS-CoV-2 genomic sequences collected in the Chiaravalle patients. Multilineage phylogenetic tree reconstructed using 913 SARS-CoV-2 sequences downloaded from GISAID database. The SARS-CoV-2 consensus genomes of the 60 Chiaravalle patients are indicated with a red dot. Inclusion of all 60 consensus genomes led to a cluster of genomes around the founder genome. The reliability of the phylogenetic clustering was evaluated using bootstrap analysis with 1000 replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)