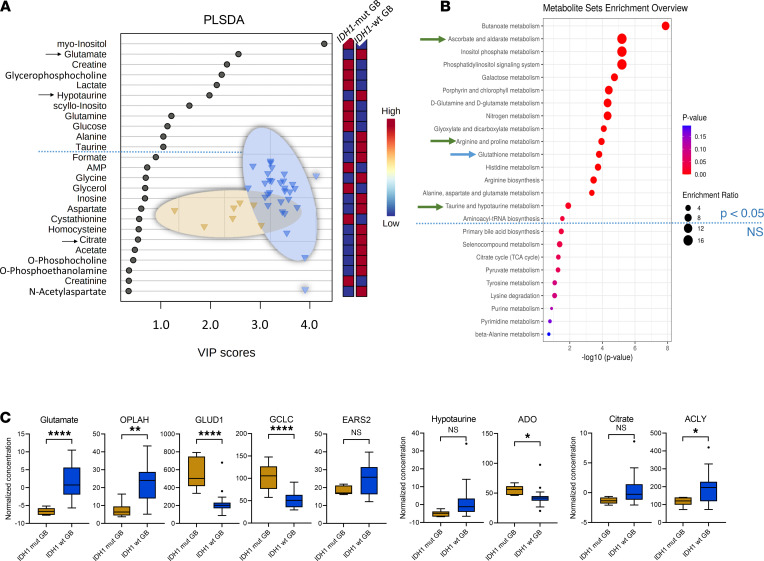
Figure 4. IDH1 mutation status cohort comparison III.
(A–C) PLS-DA identifies 11 metabolites with VIP scores > 1 (A) and 16 altered pathways (B) for comparing IDH1-mut (n = 7) versus WT (n = 30) glioblastoma (GB) subgroup (comparison III), selection of significant metabolites (as indicated by arrow in the VIP scores plot), and their corresponding gene expression changes (C) in the IDH1 mutation status cohort comparison (III) illustrated as box-and-whisker plot with Tukey’s range test. ****P < 0.0001, **P < 0.01, *P < 0.05, determined by unpaired parametric 2-tailed t test with Welch’s correction. Arrows in VIP scores plots indicate metabolites that were selected and correlated with the RNA-Seq data. Blue dotted line indicates the PLS-DA VIP score 1.0 threshold cut-off. Arrows in metabolite set enrichment overview represent most interesting and relevant metabolic pathway changes. OPLAH, 5-oxoprolinase ATP-hydrolyzing; GLUD1, glutamate dehydrogenase 1; GCLC, glutamate-cysteine ligase catalytic subunit; EARS2, mitochondrial glutamyl-TRNA synthetase 2; GOT1, glutamic-oxaloacetic transaminase 1; ACLY, ATP citrate lyase; ADO, 2-aminoethanethiol dioxygenase.