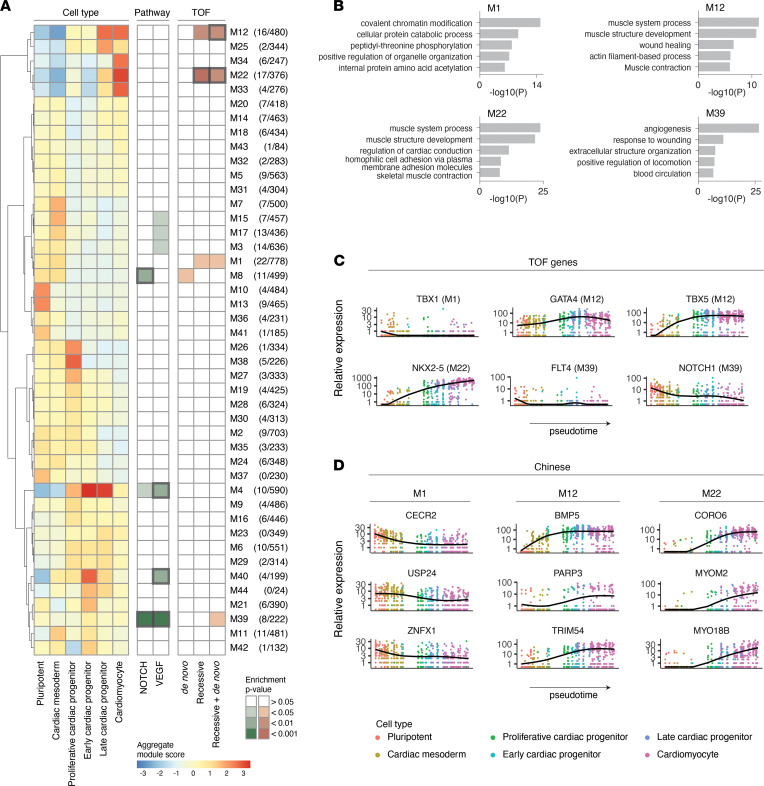
Figure 6. Integration of coexpression modules and signaling pathway enrichment with pseudotime gene expression of genes with de novo and recessive damaging variants in Chinese TOF cases.
(A) Genes varying across the in vitro cardiac differentiation trajectory grouped into 44 Monocle 3 coexpression modules and visualized as aggregate module scores for developmental states. Some modules exhibited differentiation state-specific aggregation (M10, M13, pluripotency; M38, proliferative cardiac progenitor; M22, M33, cardiac myocyte), with other modules specifically enriched for genes belonging to NOTCH (M8), VEGF (M4, M40), or both NOTCH and VEGF (M39) signaling pathways. In addition, indicated modules were enriched for genes with de novo and recessive damaging variants identified in Chinese TOF cases. The number of genes with damaging variants (total of de novo and recessive) and the total number of genes in each module are shown in parentheses. The bold rectangles in the 2 right panels indicate significance for Benjamini-Hochberg–adjusted P value at FDR < 0.10. (B) GO analysis of 4 coexpression modules showing enrichment of genes with damaging variants. The top 5 most significant GO terms are shown for each module. (C) Pseudotime expression trends of TOF genes over the pseudotime trajectory. The TOF genes are compiled from Morgenthau and Frishman (5), Page et al. (7), and Jin et al. (6). Genes belonging to the modules with enrichment of Chinese TOF candidate genes are shown, and the coexpression modules are indicated in parentheses. (D) Pseudotime expression patterns of genes with damaging de novo (CECR2, BMP5, TRIM54, and CORO6) and recessive (USP24, ZNFX1, PARP3, MYOM2, and MYO18B) mutations in Chinese TOF cases. Genes belonging to the coexpression modules with overall enrichment of damaging variants and differentiation stage-specific patterns, e.g., M1, M12, and M22, are shown.